Systems-level comparison of host-responses elicited by avian H5N1 and seasonal H1N1 influenza viruses in primary human macrophages
- PMID: 20011590
- PMCID: PMC2788213
- DOI: 10.1371/journal.pone.0008072
Systems-level comparison of host-responses elicited by avian H5N1 and seasonal H1N1 influenza viruses in primary human macrophages
Abstract
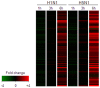
Human disease caused by highly pathogenic avian influenza (HPAI) H5N1 can lead to a rapidly progressive viral pneumonia leading to acute respiratory distress syndrome. There is increasing evidence from clinical, animal models and in vitro data, which suggests a role for virus-induced cytokine dysregulation in contributing to the pathogenesis of human H5N1 disease. The key target cells for the virus in the lung are the alveolar epithelium and alveolar macrophages, and we have shown that, compared to seasonal human influenza viruses, equivalent infecting doses of H5N1 viruses markedly up-regulate pro-inflammatory cytokines in both primary cell types in vitro. Whether this H5N1-induced dysregulation of host responses is driven by qualitative (i.e activation of unique host pathways in response to H5N1) or quantitative differences between seasonal influenza viruses is unclear. Here we used microarrays to analyze and compare the gene expression profiles in primary human macrophages at 1, 3, and 6 h after infection with H5N1 virus or low-pathogenic seasonal influenza A (H1N1) virus. We found that host responses to both viruses are qualitatively similar with the activation of nearly identical biological processes and pathways. However, in comparison to seasonal H1N1 virus, H5N1 infection elicits a quantitatively stronger host inflammatory response including type I interferon (IFN) and tumor necrosis factor (TNF)-alpha genes. A network-based analysis suggests that the synergy between IFN-beta and TNF-alpha results in an enhanced and sustained IFN and pro-inflammatory cytokine response at the early stage of viral infection that may contribute to the viral pathogenesis and this is of relevance to the design of novel therapeutic strategies for H5N1 induced respiratory disease.
Conflict of interest statement
Figures
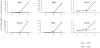

References
-
- Michaelis M, Doerr HW, Cinatl J., Jr Novel swine-origin influenza A virus in humans: another pandemic knocking at the door. Med Microbiol Immunol 2009 - PubMed
-
- Smith GJ, Vijaykrishna D, Bahl J, Lycett SJ, Worobey M, et al. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature. 2009;459:1122–1125. - PubMed
-
- Koen J. A practical method for field diagnoses of swine diseases. Am J Vet Med. 1919;14:468–470.
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

