RAD59 and RAD1 cooperate in translocation formation by single-strand annealing in Saccharomyces cerevisiae
- PMID: 20012294
- PMCID: PMC2808509
- DOI: 10.1007/s00294-009-0282-6
RAD59 and RAD1 cooperate in translocation formation by single-strand annealing in Saccharomyces cerevisiae
Abstract
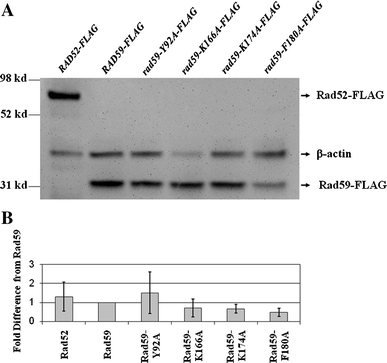
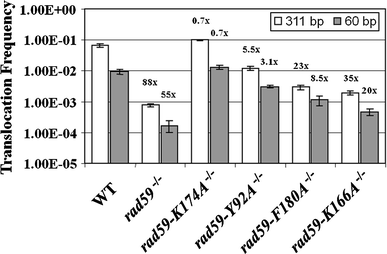
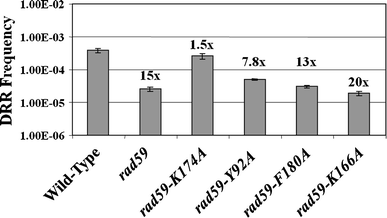
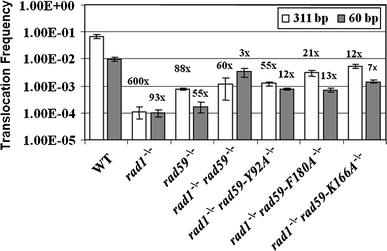
Studies in the budding yeast, Saccharomyces cerevisiae, have demonstrated that a substantial fraction of double-strand break repair following acute radiation exposure involves homologous recombination between repetitive genomic elements. We have previously described an assay in S. cerevisiae that allows us to model how repair of multiple breaks leads to the formation of chromosomal translocations by single-strand annealing (SSA) and found that Rad59, a paralog of the single-stranded DNA annealing protein Rad52, is critically important in this process. We have constructed several rad59 missense alleles to study its function more closely. Characterization of these mutants revealed proportional defects in both translocation formation and spontaneous direct-repeat recombination, which is also thought to occur by SSA. Combining the rad59 missense alleles with a null allele of RAD1, which encodes a subunit of a nuclease required for the removal of non-homologous tails from annealed intermediates, substantially suppressed the low frequency of translocations observed in rad1-null single mutants. These data suggest that at least one role of Rad59 in translocation formation by SSA is supporting the machinery required for cleavage of non-homologous tails.
Figures





Similar articles
-
Alleles of the homologous recombination gene, RAD59, identify multiple responses to disrupted DNA replication in Saccharomyces cerevisiae.BMC Microbiol. 2013 Oct 14;13:229. doi: 10.1186/1471-2180-13-229. BMC Microbiol. 2013. PMID: 24125552 Free PMC article.
-
Msh2 blocks an alternative mechanism for non-homologous tail removal during single-strand annealing in Saccharomyces cerevisiae.PLoS One. 2009 Oct 16;4(10):e7488. doi: 10.1371/journal.pone.0007488. PLoS One. 2009. PMID: 19834615 Free PMC article.
-
RAD59 is required for efficient repair of simultaneous double-strand breaks resulting in translocations in Saccharomyces cerevisiae.DNA Repair (Amst). 2008 May 3;7(5):788-800. doi: 10.1016/j.dnarep.2008.02.003. Epub 2008 Mar 25. DNA Repair (Amst). 2008. PMID: 18373960 Free PMC article.
-
Regulation of Single-Strand Annealing and its Role in Genome Maintenance.Trends Genet. 2016 Sep;32(9):566-575. doi: 10.1016/j.tig.2016.06.007. Epub 2016 Jul 19. Trends Genet. 2016. PMID: 27450436 Free PMC article. Review.
-
Emerging non-canonical roles for the Rad51-Rad52 interaction in response to double-strand breaks in yeast.Curr Genet. 2020 Oct;66(5):917-926. doi: 10.1007/s00294-020-01081-z. Epub 2020 May 12. Curr Genet. 2020. PMID: 32399607 Free PMC article. Review.
Cited by
-
Mechanism for inverted-repeat recombination induced by a replication fork barrier.Nat Commun. 2022 Jan 10;13(1):32. doi: 10.1038/s41467-021-27443-w. Nat Commun. 2022. PMID: 35013185 Free PMC article.
-
Rad59 regulates association of Rad52 with DNA double-strand breaks.Microbiologyopen. 2012 Sep;1(3):285-97. doi: 10.1002/mbo3.31. Epub 2012 Aug 3. Microbiologyopen. 2012. PMID: 23170228 Free PMC article.
-
Alleles of the homologous recombination gene, RAD59, identify multiple responses to disrupted DNA replication in Saccharomyces cerevisiae.BMC Microbiol. 2013 Oct 14;13:229. doi: 10.1186/1471-2180-13-229. BMC Microbiol. 2013. PMID: 24125552 Free PMC article.
-
Quantitation and analysis of the formation of HO-endonuclease stimulated chromosomal translocations by single-strand annealing in Saccharomyces cerevisiae.J Vis Exp. 2011 Sep 23;(55):3150. doi: 10.3791/3150. J Vis Exp. 2011. PMID: 21968396 Free PMC article.
-
Variants of the human RAD52 gene confer defects in ionizing radiation resistance and homologous recombination repair in budding yeast.Microb Cell. 2020 Jul 20;7(10):270-285. doi: 10.15698/mic2020.10.732. Microb Cell. 2020. PMID: 33015141 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

