Gene expression pathway analysis to predict response to neoadjuvant docetaxel and capecitabine for breast cancer
- PMID: 20012355
- PMCID: PMC5892182
- DOI: 10.1007/s10549-009-0651-3
Gene expression pathway analysis to predict response to neoadjuvant docetaxel and capecitabine for breast cancer
Abstract
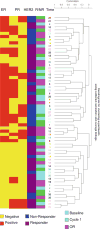
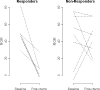
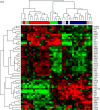
Neoadjuvant chemotherapy has been shown to be equivalent to post-operative treatment for breast cancer, and allows for assessment of chemotherapy response. In a pilot trial of docetaxel (T) and capecitabine (X) neoadjuvant chemotherapy for Stage II/III BC, we assessed correlation between baseline gene expression and tumor response to treatment, and examined changes in gene expression associated with treatment. Patients received four cycles of TX. Tumor tissue obtained from Mammotome core biopsies pretreatment (BL) and post-cycle 1 (C1) of TX was FLash frozen and stored at -70 degrees C until processing. Gene expression analysis utilized Affymetrix HG-U133 Plus 2.0 GeneChip arrays. Statistical analysis was performed using BRB Array Tools after RMA normalization. Gene ontology (GO) pathway analysis used random variance t tests with a significance level of P\0.005. For gene categories identified byGO pathway analysis as significant, expression levels of individual genes within those pathways were compared between classes using univariate t tests; those genes with significance level of P\0.05 were reported. PAM50 analyses were performed on tumor samples to investigate biologic subtype and risk of relapse (ROR). Using GO pathway analysis, 39 gene categories discriminated between responders and non-responders,most notably genes involved in microtubule assembly and regulation. When comparing pre- and post-chemotherapy specimens, we identified 71 differentially expressed gene categories, including DNA repair and cell proliferation regulation. There were 45 GO pathways in which the change in expression after one cycle of chemotherapy was significantly different among responders and nonresponders. The majority of tumor samples fell into the basal like and luminal B categories. ROR scores decreased in response to chemotherapy; this change was more evident in samples from patients classified as responders by clinical criteria. GO pathway analysis identified a number of gene categories pertinent to therapeutic response, and may be an informative method for identifying genes important in response to chemotherapy. Larger studies using the methods described here are necessary to fully evaluate gene expression changes in response to chemotherapy.
Figures

References
-
- Berry DA, Cronin KA, Plevritis SK, Fryback DG, Clarke L, Zelen M, et al. Effect of screening and adjuvant therapy on mortality from breast cancer. N Engl J Med. 2005;353(17):1784–1792. - PubMed
-
- Harlan LC, Clegg LX, Abrams J, Stevens JL, Ballard-Barbash R. Community-based use of chemotherapy and hormonal therapy for early-stage breast cancer: 1987–2000. J Clin Oncol. 2006;24(6):872–877. - PubMed
-
- Andre F, Mazouni C, Hortobagyi GN, Pusztai L. DNA arrays as predictors of efficacy of adjuvant/neoadjuvant chemotherapy in breast cancer patients: current data and issues on study design. Biochim Biophys Acta. 2006;1766(2):197–204. - PubMed
-
- Chuthapisith S, Eremin JM, Eremin O. Predicting response to neoadjuvant chemotherapy in breast cancer: molecular imaging, systemic biomarkers and the cancer metabolome (review) Oncol Rep. 2008;20(4):699–703. - PubMed
-
- Lebowitz PF, Eng-Wong J, Swain SM, Berman A, Merino MJ, Chow CK, et al. A phase II trial of neoadjuvant docetaxel and capecitabine for locally advanced breast cancer. Clin Cancer Res. 2004;10(20):6764–6769. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

