The alpha-kinase family: an exceptional branch on the protein kinase tree
- PMID: 20012461
- PMCID: PMC2827801
- DOI: 10.1007/s00018-009-0215-z
The alpha-kinase family: an exceptional branch on the protein kinase tree
Abstract
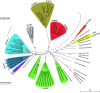
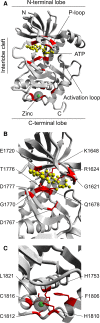
The alpha-kinase family represents a class of atypical protein kinases that display little sequence similarity to conventional protein kinases. Early studies on myosin heavy chain kinases in Dictyostelium discoideum revealed their unusual propensity to phosphorylate serine and threonine residues in the context of an alpha-helix. Although recent studies show that some members of this family can also phosphorylate residues in non-helical regions, the name alpha-kinase has remained. During evolution, the alpha-kinase domains combined with many different functional subdomains such as von Willebrand factor-like motifs (vWKa) and even cation channels (TRPM6 and TRPM7). As a result, these kinases are implicated in a large variety of cellular processes such as protein translation, Mg(2+) homeostasis, intracellular transport, cell migration, adhesion, and proliferation. Here, we review the current state of knowledge on different members of this kinase family and discuss the potential use of alpha-kinases as drug targets in diseases such as cancer.
Figures

References
-
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. - PubMed
-
- Drennan D, Ryazanov AG. Alpha-kinases: analysis of the family and comparison with conventional protein kinases. Prog Biophys Mol Biol. 2004;85:1–32. - PubMed
-
- Ryazanov AG, Pavur KS, Dorovkov MV. Alpha-kinases: a new class of protein kinases with a novel catalytic domain. Curr Biol. 1999;9:R43–R45. - PubMed
-
- Futey LM, Medley QG, Cote GP, Egelhoff TT. Structural analysis of myosin heavy chain kinase A from Dictyostelium. Evidence for a highly divergent protein kinase domain, an amino-terminal coiled-coil domain, and a domain homologous to the beta-subunit of heterotrimeric G proteins. J Biol Chem. 1995;270:523–529. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

