Genetic and biochemical analysis of the TLA1 gene in Chlamydomonas reinhardtii
- PMID: 20012986
- PMCID: PMC2806527
- DOI: 10.1007/s00425-009-1083-3
Genetic and biochemical analysis of the TLA1 gene in Chlamydomonas reinhardtii
Abstract
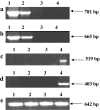
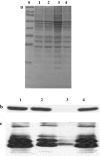
The Chlamydomonas reinhardtii genomic DNA database contains a predicted open reading frame (ORF-P) without an apparent stop-codon and unknown coding sequence, located in close proximity and immediately upstream of the TLA1 gene (GenBank Accession No. AF534570). The latter was implicated in the regulation of the light-harvesting chlorophyll antenna size of photosynthesis (Tetali et al. Planta 225:813-829, 2007). To provide currently lacking information on ORF-P and its potential participation in TLA1 gene expression, thus in the regulation of the chlorophyll antenna size, genetic and biochemical analyses were undertaken. The coding and UTR regions of the ORF-P were defined and delineated from those of the adjacent TLA1 gene. ORF-P is shown to encode a protein with a distinct RING-like zinc finger domain that is present in numerous eukaryotic proteins, believed to play a role in cellular ubiquitination, leading to regulation of cellular processes like signaling, growth, transcription, and DNA repair. It is further shown that the two genes share a 74-bp overlap between the 3' UTR region of ORF-P and the 5' UTR region of TLA1. However, they possess distinct start and stop codons and separate coding sequences, and transcribed as separate mRNAs without any trans-splicing between them. Complementation experiments showed that the TLA1 gene alone is sufficient to rescue the truncated chlorophyll antenna size phenotype of the tla1 mutant. Protein sequence alignments in C. reinhardtii and the colorless microalga Polytomella parva suggested that TLA1 defines the relationship between nucleus and organelle in microalgae, indirectly affecting the development of the chlorophyll antenna size.
Figures





Similar articles
-
Polyclonal antibodies against the TLA1 protein also recognize with high specificity the D2 reaction center protein of PSII in the green alga Chlamydomonas reinhardtii.Photosynth Res. 2012 Apr;112(1):39-47. doi: 10.1007/s11120-012-9733-x. Epub 2012 Mar 23. Photosynth Res. 2012. PMID: 22442055
-
tla1, a DNA insertional transformant of the green alga Chlamydomonas reinhardtii with a truncated light-harvesting chlorophyll antenna size.Planta. 2003 May;217(1):49-59. doi: 10.1007/s00425-002-0968-1. Epub 2003 Feb 12. Planta. 2003. PMID: 12721848
-
Development of the light-harvesting chlorophyll antenna in the green alga Chlamydomonas reinhardtii is regulated by the novel Tla1 gene.Planta. 2007 Mar;225(4):813-29. doi: 10.1007/s00425-006-0392-z. Planta. 2007. PMID: 16977454
-
A genome's-eye view of the light-harvesting polypeptides of Chlamydomonas reinhardtii.Curr Genet. 2004 Feb;45(2):61-75. doi: 10.1007/s00294-003-0460-x. Epub 2003 Dec 2. Curr Genet. 2004. PMID: 14652691 Review.
-
Novel insights into the function of LHCSR3 in Chlamydomonas reinhardtii.Plant Signal Behav. 2015;10(12):e1058462. doi: 10.1080/15592324.2015.1058462. Plant Signal Behav. 2015. PMID: 26237677 Free PMC article. Review.
Cited by
-
Assembly of the light-harvesting chlorophyll antenna in the green alga Chlamydomonas reinhardtii requires expression of the TLA2-CpFTSY gene.Plant Physiol. 2012 Feb;158(2):930-45. doi: 10.1104/pp.111.189910. Epub 2011 Nov 23. Plant Physiol. 2012. PMID: 22114096 Free PMC article.
-
Polyclonal antibodies against the TLA1 protein also recognize with high specificity the D2 reaction center protein of PSII in the green alga Chlamydomonas reinhardtii.Photosynth Res. 2012 Apr;112(1):39-47. doi: 10.1007/s11120-012-9733-x. Epub 2012 Mar 23. Photosynth Res. 2012. PMID: 22442055
-
Engineering photosynthetic organisms for the production of biohydrogen.Photosynth Res. 2015 Mar;123(3):241-53. doi: 10.1007/s11120-014-9991-x. Epub 2014 Mar 27. Photosynth Res. 2015. PMID: 24671643 Free PMC article. Review.
-
Optimizing antenna size to maximize photosynthetic efficiency.Plant Physiol. 2011 Jan;155(1):79-85. doi: 10.1104/pp.110.165886. Epub 2010 Nov 15. Plant Physiol. 2011. PMID: 21078863 Free PMC article. No abstract available.
-
Modulation of the light-harvesting chlorophyll antenna size in Chlamydomonas reinhardtii by TLA1 gene over-expression and RNA interference.Philos Trans R Soc Lond B Biol Sci. 2012 Dec 19;367(1608):3430-43. doi: 10.1098/rstb.2012.0229. Philos Trans R Soc Lond B Biol Sci. 2012. PMID: 23148270 Free PMC article.
References
-
- Anderson JM. Photoregulation of the composition, function and structure of thylakoid membranes. Annu Rev Plant Physiol. 1986;37:93–136. doi: 10.1146/annurev.pp.37.060186.000521. - DOI
-
- Bjorkman O, Boardman NK, Anderson JM, Thorne SW, Goodchild DJ, Pyliotis NA (1972) Effect of light intensity during growth of Atriplex patula on the capacity of photosynthetic reactions, chloroplast components and structure. Carnegie Inst Yearbook 71:115–135
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

