Multiple base-recognition sites in a biological nanopore: two heads are better than one
- PMID: 20014084
- PMCID: PMC3128935
- DOI: 10.1002/anie.200905483
Multiple base-recognition sites in a biological nanopore: two heads are better than one
Abstract
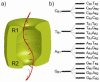
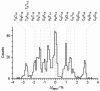
Ultra-rapid sequencing of DNA strands with nanopores is under intense investigation. The αHL protein nanopore is a leading candidate sensor for this approach. Multiple base-recognition sites have been identified in engineered αHL pores. By using immobilized synthetic oligonucleotides, we show here that additional sequence information can be gained when two recognition sites, rather than one, are employed within a single nanopore.
Figures


Similar articles
-
Nanopore sequencing technology: nanopore preparations.Trends Biotechnol. 2007 Apr;25(4):174-81. doi: 10.1016/j.tibtech.2007.02.008. Epub 2007 Feb 22. Trends Biotechnol. 2007. PMID: 17320228 Review.
-
Urea facilitates the translocation of single-stranded DNA and RNA through the alpha-hemolysin nanopore.Biophys J. 2010 May 19;98(9):1856-63. doi: 10.1016/j.bpj.2009.12.4333. Biophys J. 2010. PMID: 20441749 Free PMC article.
-
Nanopore sequencing: from imagination to reality.Clin Chem. 2015 Jan;61(1):25-31. doi: 10.1373/clinchem.2014.223016. Epub 2014 Dec 4. Clin Chem. 2015. PMID: 25477535 Free PMC article. No abstract available.
-
Control and reversal of the electrophoretic force on DNA in a charged nanopore.J Phys Condens Matter. 2010 Nov 17;22(45):454123. doi: 10.1088/0953-8984/22/45/454123. Epub 2010 Oct 29. J Phys Condens Matter. 2010. PMID: 21339610 Free PMC article.
-
Peering into biological nanopore: a practical technology to single-molecule analysis.Chem Asian J. 2010 Sep 3;5(9):1952-61. doi: 10.1002/asia.201000279. Chem Asian J. 2010. PMID: 20669216 Review.
Cited by
-
How capture affects polymer translocation in a solitary nanopore.J Chem Phys. 2022 Jun 28;156(24):244902. doi: 10.1063/5.0094221. J Chem Phys. 2022. PMID: 35778106 Free PMC article.
-
Nucleobase recognition in ssDNA at the central constriction of the alpha-hemolysin pore.Nano Lett. 2010 Sep 8;10(9):3633-7. doi: 10.1021/nl101955a. Nano Lett. 2010. PMID: 20704324 Free PMC article.
-
A Protein Rotaxane Controls the Translocation of Proteins Across a ClyA Nanopore.Nano Lett. 2015 Sep 9;15(9):6076-6081. doi: 10.1021/acs.nanolett.5b02309. Epub 2015 Aug 7. Nano Lett. 2015. PMID: 26243210 Free PMC article.
-
Recent advances in biological nanopores for nanopore sequencing, sensing and comparison of functional variations in MspA mutants.RSC Adv. 2021 Aug 31;11(46):28996-29014. doi: 10.1039/d1ra02364k. eCollection 2021 Aug 23. RSC Adv. 2021. PMID: 35478559 Free PMC article. Review.
-
Giant single molecule chemistry events observed from a tetrachloroaurate(III) embedded Mycobacterium smegmatis porin A nanopore.Nat Commun. 2019 Dec 11;10(1):5668. doi: 10.1038/s41467-019-13677-2. Nat Commun. 2019. PMID: 31827098 Free PMC article.
References
-
- Branton D, Deamer DW, Marziali A, Bayley H, Benner SA, Butler T, Di Ventra M, Garaj S, Hibbs A, Huang X, Jovanovich SB, Krstic PS, Lindsay S, Ling XS, Mastrangelo CH, Meller A, Oliver JS, Pershin YV, Ramsey JM, Riehn R, Soni GV, Tabard-Cossa V, Wanunu M, Wiggin M, Schloss JA. Nature Biotechnology. 2008;26:1146. - PMC - PubMed
-
- Purnell RF, Schmidt JJ. ACS Nano. 2009 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

