Multiple base-recognition sites in a biological nanopore: two heads are better than one
- PMID: 20014084
- PMCID: PMC3128935
- DOI: 10.1002/anie.200905483
Multiple base-recognition sites in a biological nanopore: two heads are better than one
Abstract
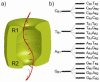
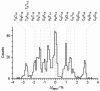
Ultra-rapid sequencing of DNA strands with nanopores is under intense investigation. The αHL protein nanopore is a leading candidate sensor for this approach. Multiple base-recognition sites have been identified in engineered αHL pores. By using immobilized synthetic oligonucleotides, we show here that additional sequence information can be gained when two recognition sites, rather than one, are employed within a single nanopore.
Figures


References
-
- Branton D, Deamer DW, Marziali A, Bayley H, Benner SA, Butler T, Di Ventra M, Garaj S, Hibbs A, Huang X, Jovanovich SB, Krstic PS, Lindsay S, Ling XS, Mastrangelo CH, Meller A, Oliver JS, Pershin YV, Ramsey JM, Riehn R, Soni GV, Tabard-Cossa V, Wanunu M, Wiggin M, Schloss JA. Nature Biotechnology. 2008;26:1146. - PMC - PubMed
-
- Purnell RF, Schmidt JJ. ACS Nano. 2009 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

