RAGE-independent autoreactive B cell activation in response to chromatin and HMGB1/DNA immune complexes
- PMID: 20014975
- PMCID: PMC2929824
- DOI: 10.3109/08916930903384591
RAGE-independent autoreactive B cell activation in response to chromatin and HMGB1/DNA immune complexes
Abstract
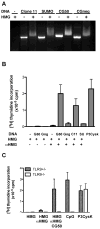
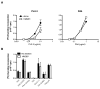
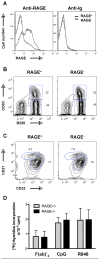
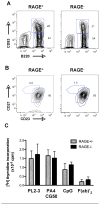
Increasing evidence suggests that the excessive accumulation of apoptotic or necrotic cellular debris may contribute to the pathology of systemic autoimmune disease. HMGB1 is a nuclear DNA-associated protein, which can be released from dying cells thereby triggering inflammatory processes. We have previously shown that IgG2a-reactive B cell receptor (BCR) transgenic AM14 B cells proliferate in response to endogenous chromatin immune complexes (ICs), in the form of the anti-nucleosome antibody PL2-3 and cell debris, in a TLR9-dependent manner, and that these ICs contain HMGB1. Activation of AM14 B cells by these chromatin ICs was inhibited by a soluble form of the HMGB1 receptor, RAGE-Fc, suggesting HMGB1-RAGE interaction was important for this response. To further explore the role of HMGB1 in autoreactive B cell activation, we assessed the capacity of purified calf thymus HMGB1 to bind dsDNA fragments and found that HMGB1 bound both CG-rich and CG-poor DNA. However, HMGB1-DNA complexes could not activate AM14 B cells unless HMGB1 was bound by IgG2a and thereby able to engage the BCR. To ascertain the role of RAGE in autoreactive B cell responses to chromatin ICs, we intercrossed AM14 and RAGE-deficient mice. We found that spontaneous and defined DNA ICs activated RAGE+ and RAGE(- ) AM14 B cells to a comparable extent. These results suggest that HMGB1 promotes B cell responses to endogenous TLR9 ligands through a RAGE-independent mechanism.
Figures
References
-
- Tian J, Avalos AM, Mao SY, Chen B, Senthil K, Wu H, Parroche P, Drabic S, Golenbock D, Sirois C, et al. Toll-like receptor 9-dependent activation by DNA-containing immune complexes is mediated by HMGB1 and RAGE. Nat Immunol. 2007;8:487–96. - PubMed
-
- Bianchi ME, Manfredi A. Chromatin and cell death. Biochim Biophys Acta. 2004;1677:181–6. - PubMed
-
- Thomas JO. HMG1 and 2: architectural DNA-binding proteins. Biochem Soc Trans. 2001;29:395–401. - PubMed
-
- Scaffidi P, Misteli T, Bianchi ME. Release of chromatin protein HMGB1 by necrotic cells triggers inflammation. Nature. 2002;418:191–5. - PubMed
-
- Bell CW, Jiang W, Reich CF, 3rd, Pisetsky DS. The extracellular release of HMGB1 during apoptotic cell death. Am J Physiol Cell Physiol. 2006;291:C1318–25. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
