Relaxin gene family in teleosts: phylogeny, syntenic mapping, selective constraint, and expression analysis
- PMID: 20015397
- PMCID: PMC2805637
- DOI: 10.1186/1471-2148-9-293
Relaxin gene family in teleosts: phylogeny, syntenic mapping, selective constraint, and expression analysis
Abstract
Background: In recent years, the relaxin family of signaling molecules has been shown to play diverse roles in mammalian physiology, but little is known about its diversity or physiology in teleosts, an infraclass of the bony fishes comprising approximately 50% of all extant vertebrates. In this paper, 32 relaxin family sequences were obtained by searching genomic and cDNA databases from eight teleost species; phylogenetic, molecular evolutionary, and syntenic data analyses were conducted to understand the relationship and differential patterns of evolution of relaxin family genes in teleosts compared with mammals. Additionally, real-time quantitative PCR was used to confirm and assess the tissues of expression of five relaxin family genes in Danio rerio and in situ hybridization used to assess the site-specific expression of the insulin 3-like gene in D. rerio testis.
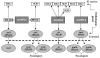
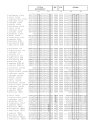
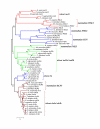
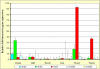
Results: Up to six relaxin family genes were identified in each teleost species. Comparative syntenic mapping revealed that fish possess two paralogous copies of human RLN3, which we call rln3a and rln3b, an orthologue of human RLN2, rln, two paralogous copies of human INSL5, insl5a and insl5b, and an orthologue of human INSL3, insl3. Molecular evolutionary analyses indicated that: rln3a, rln3b and rln are under strong evolutionary constraint, that insl3 has been subject to moderate rates of sequence evolution with two amino acids in insl3/INSL3 showing evidence of positively selection, and that insl5b exhibits a higher rate of sequence evolution than its paralogue insl5a suggesting that it may have been neo-functionalized after the teleost whole genome duplication. Quantitative PCR analyses in D. rerio indicated that rln3a and rln3b are expressed in brain, insl3 is highly expressed in gonads, and that there was low expression of both insl5 genes in adult zebrafish. Finally, in situ hybridization of insl3 in D. rerio testes showed highly specific hybridization to interstitial Leydig cells.
Conclusions: Contrary to previous studies, we find convincing evidence that teleosts contain orthologues of four relaxin family peptides. Overall our analyses suggest that in teleosts: 1) rln3 exhibits a similar evolution and expression pattern to mammalian RLN3, 2) insl3 has been subject to positive selection like its mammalian counterpart and shows similar tissue-specific expression in Leydig cells, 3) insl5 genes are highly represented and have a relatively high rate of sequence evolution in teleost genomes, but they exhibited only low levels of expression in adult zebrafish, 4) rln is evolving under very different selective constraints from mammalian RLN. The results presented here should facilitate the development of hypothesis-driven experimental work on the specific roles of relaxin family genes in teleosts.
Figures



Similar articles
-
Temporal and spatial expression of insulin-like peptide (insl5a and insl5b) paralog genes during the embryogenesis of Danio rerio.J Exp Zool B Mol Dev Evol. 2018 Jan;330(1):33-40. doi: 10.1002/jez.b.22787. Epub 2018 Jan 10. J Exp Zool B Mol Dev Evol. 2018. PMID: 29319231
-
Relaxin family genes in humans and teleosts.Ann N Y Acad Sci. 2009 Apr;1160:42-4. doi: 10.1111/j.1749-6632.2009.03842.x. Ann N Y Acad Sci. 2009. PMID: 19416157
-
Japanese medaka as a model for studying the relaxin family genes involved in neuroendocrine regulation: Insights from the expression of fish-specific rln3 and insl5 and rxfp3/4-type receptor paralogues.Mol Cell Endocrinol. 2019 May 1;487:2-11. doi: 10.1016/j.mce.2019.01.017. Epub 2019 Jan 28. Mol Cell Endocrinol. 2019. PMID: 30703485
-
The relaxin family peptide receptors and their ligands: new developments and paradigms in the evolution from jawless fish to mammals.Gen Comp Endocrinol. 2014 Dec 1;209:93-105. doi: 10.1016/j.ygcen.2014.07.014. Epub 2014 Jul 29. Gen Comp Endocrinol. 2014. PMID: 25079565 Review.
-
Relaxin family peptides in the male reproductive system--a critical appraisal.Mol Hum Reprod. 2011 Feb;17(2):71-84. doi: 10.1093/molehr/gaq086. Epub 2010 Oct 14. Mol Hum Reprod. 2011. PMID: 20952422 Review.
Cited by
-
New insights into ligand-receptor pairing and coevolution of relaxin family peptides and their receptors in teleosts.Int J Evol Biol. 2012;2012:310278. doi: 10.1155/2012/310278. Epub 2012 Sep 13. Int J Evol Biol. 2012. PMID: 23008798 Free PMC article.
-
Insulin-like 3 affects zebrafish spermatogenic cells directly and via Sertoli cells.Commun Biol. 2021 Feb 15;4(1):204. doi: 10.1038/s42003-021-01708-y. Commun Biol. 2021. PMID: 33589679 Free PMC article.
-
Distinct and Cooperative Roles of amh and dmrt1 in Self-Renewal and Differentiation of Male Germ Cells in Zebrafish.Genetics. 2017 Nov;207(3):1007-1022. doi: 10.1534/genetics.117.300274. Epub 2017 Sep 11. Genetics. 2017. PMID: 28893856 Free PMC article.
-
Comparative Genomic Characterization of Relaxin Peptide Family in Cattle and Buffalo.Biomed Res Int. 2022 Oct 4;2022:1581714. doi: 10.1155/2022/1581714. eCollection 2022. Biomed Res Int. 2022. PMID: 36246983 Free PMC article.
-
Characterization of the prohormone complement in Amphiprion and related fish species integrating genome and transcriptome assemblies.PLoS One. 2020 Mar 12;15(3):e0228562. doi: 10.1371/journal.pone.0228562. eCollection 2020. PLoS One. 2020. PMID: 32163422 Free PMC article.
References
-
- Hsu SY, Kudo M, Chen T, Nakabayashi K, Bhalla A, Spek PJ van der, van Duin M, Hsueh AJ. The three subfamilies of leucine-rich repeat-containing G protein-coupled receptors (LGR): identification of LGR6 and LGR7 and the signaling mechanism for LGR7. Mol Endocrinol. 2000;14:1257–1271. doi: 10.1210/me.14.8.1257. - DOI - PubMed
-
- Sherwood OD. In: The physiology of reproduction. 2. Knobil E, Neill JD, editor. Vol. 1. New York: Raven Press; 1994. Relaxin; pp. 861–1009.
-
- McGuane JT, Parry LJ. Relaxin and the extracellular matrix: molecular mechanisms of action and implications for cardiovascular disease. Expert Rev Mol Medicine. 2005;7:1–18. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

