The Bmi-1 polycomb protein antagonizes the (-)-epigallocatechin-3-gallate-dependent suppression of skin cancer cell survival
- PMID: 20015867
- PMCID: PMC2832547
- DOI: 10.1093/carcin/bgp314
The Bmi-1 polycomb protein antagonizes the (-)-epigallocatechin-3-gallate-dependent suppression of skin cancer cell survival
Abstract
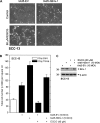
The polycomb group (PcG) proteins are epigenetic regulators of gene expression that enhance cell survival. This regulation is achieved via action of two multiprotein PcG complexes--PRC2 (EED) and PRC1 [B-cell-specific Moloney murine leukemia virus integration site 1 (Bmi-1)]. These complexes modulate gene expression by increasing histone methylation and reducing acetylation--leading to a closed chromatin conformation. Activity of these proteins is associated with increased cell proliferation and survival. We show increased expression of key PcG proteins in immortalized keratinocytes and skin cancer cell lines. We examine the role of two key PcG proteins, Bmi-1 and enhancer of zeste homolog 2 (Ezh2), and the impact of the active agent in green tea, (-)-epigallocatechin-3-gallate (EGCG), on the function of these regulators. EGCG treatment of SCC-13 cells reduces Bmi-1 and Ezh2 level and this is associated with reduced cell survival. The reduction in survival is associated with a global reduction in histone H3 lysine 27 trimethylation, a hallmark of PRC2 complex action. This change in PcG protein expression is associated with reduced expression of key proteins that enhance progression through the cell cycle [cyclin-dependent kinase (cdk)1, cdk2, cdk4, cyclin D1, cyclin E, cyclin A and cyclin B1] and increased expression of proteins that inhibit cell cycle progression (p21 and p27). Apoptosis is also enhanced, as evidenced by increased caspase 9, 8 and 3 cleavage and increased poly(adenosine diphosphate ribose) polymerase cleavage. EGCG treatment also increases Bax and suppresses Bcl-xL expression. Vector-mediated enhanced Bmi-1 expression reverses these EGCG-dependent changes. These findings suggest that green tea polyphenols reduce skin tumor cell survival by influencing PcG-mediated epigenetic regulatory mechanisms.
Figures







Comment in
-
Findings of Research Misconduct.Fed Regist. 2024 Aug 15;89(158):66420-66422. Fed Regist. 2024. PMID: 39161428 Free PMC article. No abstract available.
References
-
- Sparmann A, et al. Polycomb silencers control cell fate, development and cancer. Nat. Rev. Cancer. 2006;6:846–856. - PubMed
-
- Vaissiere T, et al. Epigenetic interplay between histone modifications and DNA methylation in gene silencing. Mutat. Res. 2008;659:40–48. - PubMed
-
- Jacobs JJ, et al. Polycomb repression: from cellular memory to cellular proliferation and cancer. Biochim. Biophys. Acta. 2002;1602:151–161. - PubMed
-
- Jacobs JJ, et al. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397:164–168. - PubMed
-
- Orlando V. Polycomb, epigenomes, and control of cell identity. Cell. 2003;112:599–606. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

