Loss of IGF2 imprinting is associated with abrogation of long-range intrachromosomal interactions in human cancer cells
- PMID: 20015958
- PMCID: PMC2816615
- DOI: 10.1093/hmg/ddp558
Loss of IGF2 imprinting is associated with abrogation of long-range intrachromosomal interactions in human cancer cells
Abstract
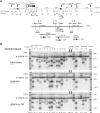
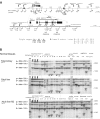
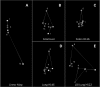
Nuclear architecture and chromatin geography are important factors in the regulation of gene expression, as these components may play a vital epigenetic role both in normal physiology as well as in the initiation and progression of malignancies. Using a modification of the chromosome conformation capture (3C) technique, we examined long-range chromatin interactions of the imprinted human IGF2 gene. We demonstrate that numerous intrachromosomal interactions occur along both parental alleles in normal tissues, where the IGF2 is paternally expressed, as well as in normal liver where gene expression is biallelic. Long-range and allele-specific interactions occur between the IGF2/H19 imprinting control region-1 (ICR1) and ICR2, a region which regulates an imprinted gene cluster nearly a megabase distant from IGF2. Loss of genomic imprinting is a common epigenetic event in cancer, and long-range interactions have not been examined in malignant cells. In cancer cell lines in which IGF2 imprinting is maintained (MOI), essentially all of the 3C interactions seen in normal cells were preserved. However, in cells in which IGF2 imprinting was lost (LOI), nearly all of the long-range chromatin interactions involving IGF2 were abrogated. A three-dimensional computer model depicts the physical interactions between the IGF2 promoter and ICR1 in MOI cells, while the model of LOI lung cancer cells is flattened with few long-range interactions. This dramatic change in the three-dimension configuration of the chromatin at the IGF2 locus in LOI cancer cells suggests that the loss of imprinting may lead to a variety of changes in gene expression in addition to changes in IGF2 transcription.
Figures







Similar articles
-
Alterations in promoter usage and expression levels of insulin-like growth factor-II and H19 genes in cervical carcinoma exhibiting biallelic expression of IGF-II.Biochim Biophys Acta. 2002 Apr 24;1586(3):307-15. doi: 10.1016/s0925-4439(01)00109-0. Biochim Biophys Acta. 2002. PMID: 11997082
-
Loss of insulin-like growth factor II imprinting is a hallmark associated with enhanced chemo/radiotherapy resistance in cancer stem cells.Oncotarget. 2016 Aug 9;7(32):51349-51364. doi: 10.18632/oncotarget.9784. Oncotarget. 2016. PMID: 27275535 Free PMC article.
-
Epigenetic modulation of the IGF2/H19 imprinted domain in human embryonic and extra-embryonic compartments and its possible role in fetal growth restriction.Epigenetics. 2010 May 16;5(4):313-24. doi: 10.4161/epi.5.4.11637. Epub 2010 May 28. Epigenetics. 2010. PMID: 20418667
-
Loss of IGF2 imprinting: mechanisms and consequences.Novartis Found Symp. 2004;262:108-21; discussion 121-4, 265-8. Novartis Found Symp. 2004. PMID: 15562825 Review.
-
Mechanisms of Igf2/H19 imprinting: DNA methylation, chromatin and long-distance gene regulation.J Biochem. 2000 May;127(5):711-5. doi: 10.1093/oxfordjournals.jbchem.a022661. J Biochem. 2000. PMID: 10788777 Review.
Cited by
-
Estrogen-mediated epigenetic repression of the imprinted gene cyclin-dependent kinase inhibitor 1C in breast cancer cells.Carcinogenesis. 2011 Jun;32(6):812-21. doi: 10.1093/carcin/bgr017. Epub 2011 Feb 8. Carcinogenesis. 2011. PMID: 21304052 Free PMC article.
-
Genomic interaction profiles in breast cancer reveal altered chromatin architecture.PLoS One. 2013 Sep 3;8(9):e73974. doi: 10.1371/journal.pone.0073974. eCollection 2013. PLoS One. 2013. PMID: 24019942 Free PMC article.
-
Specific changes in the expression of imprinted genes in prostate cancer--implications for cancer progression and epigenetic regulation.Asian J Androl. 2012 May;14(3):436-50. doi: 10.1038/aja.2011.160. Epub 2012 Feb 27. Asian J Androl. 2012. PMID: 22367183 Free PMC article. Review.
-
Loss of imprinting of IGF2 and the epigenetic progenitor model of cancer.Am J Stem Cells. 2011 Aug 19;1(1):59-74. Print 2012. Am J Stem Cells. 2011. PMID: 23671798 Free PMC article.
-
Chromatin structure, epigenetic mechanisms and long-range interactions in the human insulin locus.Diabetes Obes Metab. 2012 Oct;14 Suppl 3(Suppl 3):1-11. doi: 10.1111/j.1463-1326.2012.01645.x. Diabetes Obes Metab. 2012. PMID: 22928559 Free PMC article. Review.
References
-
- DeChiara T.M., Robertson E.J., Efstratiadis A. Parental imprinting of the mouse insulin-like growth factor II gene. Cell. 1991;64:849–859. - PubMed
-
- Bartolomei M.S., Zemel S., Tilghman S.M. Parental imprinting of the mouse H19 gene. Nature. 1991;351:153–155. - PubMed
-
- Zemel S., Bartolomei M.S., Tilghman S.M. Physical linkage of two mammalian imprinted genes, H19 and insulin-like growth factor 2. Nat. Genet. 1992;2:61–65. - PubMed
-
- Giannoukakis N., Deal C., Paquette J., Goodyer C.G., Polychronakos C. Parental genomic imprinting of the human IGF2 gene. Nat. Genet. 1993;4:98–101. - PubMed
-
- Ohlsson R., Nystrom A., Pfeifer O.S., Tohonen V., Hedborg F., Schofield P., Flam F., Ekstrom T.J. IGF2 is parentally imprinted during human embryogenesis and in the Beckwith–Wiedemann syndrome. Nat. Genet. 1993;4:94–97. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

