The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants
- PMID: 20015970
- PMCID: PMC2847217
- DOI: 10.1093/nar/gkp1137
The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants
Abstract
FASTQ has emerged as a common file format for sharing sequencing read data combining both the sequence and an associated per base quality score, despite lacking any formal definition to date, and existing in at least three incompatible variants. This article defines the FASTQ format, covering the original Sanger standard, the Solexa/Illumina variants and conversion between them, based on publicly available information such as the MAQ documentation and conventions recently agreed by the Open Bioinformatics Foundation projects Biopython, BioPerl, BioRuby, BioJava and EMBOSS. Being an open access publication, it is hoped that this description, with the example files provided as Supplementary Data, will serve in future as a reference for this important file format.
Figures
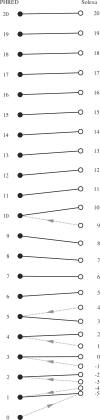
References
-
- Bennett S. Solexa Ltd. Pharmacogenomics. 2004;5:433–438. - PubMed
-
- Pandey V, Nutter RC, E EP. Applied Biosystems SOLiD system: ligation-based sequencing. In: Janitz M, editor. Next Generation Genome Sequencing: Towards Personalized Medicine. Wiley; 2008. pp. 29–41.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

