PANTHER version 7: improved phylogenetic trees, orthologs and collaboration with the Gene Ontology Consortium
- PMID: 20015972
- PMCID: PMC2808919
- DOI: 10.1093/nar/gkp1019
PANTHER version 7: improved phylogenetic trees, orthologs and collaboration with the Gene Ontology Consortium
Abstract
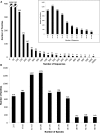
Protein Analysis THrough Evolutionary Relationships (PANTHER) is a comprehensive software system for inferring the functions of genes based on their evolutionary relationships. Phylogenetic trees of gene families form the basis for PANTHER and these trees are annotated with ontology terms describing the evolution of gene function from ancestral to modern day genes. One of the main applications of PANTHER is in accurate prediction of the functions of uncharacterized genes, based on their evolutionary relationships to genes with functions known from experiment. The PANTHER website, freely available at http://www.pantherdb.org, also includes software tools for analyzing genomic data relative to known and inferred gene functions. Since 2007, there have been several new developments to PANTHER: (i) improved phylogenetic trees, explicitly representing speciation and gene duplication events, (ii) identification of gene orthologs, including least diverged orthologs (best one-to-one pairs), (iii) coverage of more genomes (48 genomes, up to 87% of genes in each genome; see http://www.pantherdb.org/panther/summaryStats.jsp), (iv) improved support for alternative database identifiers for genes, proteins and microarray probes and (v) adoption of the SBGN standard for display of biological pathways. In addition, PANTHER trees are being annotated with gene function as part of the Gene Ontology Reference Genome project, resulting in an increasing number of curated functional annotations.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

