Genomewide association analysis of respiratory syncytial virus infection in mice
- PMID: 20015999
- PMCID: PMC2820899
- DOI: 10.1128/JVI.00584-09
Genomewide association analysis of respiratory syncytial virus infection in mice
Abstract
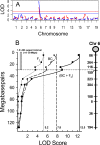
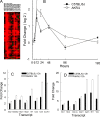
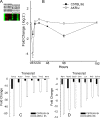
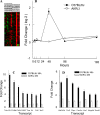
Respiratory syncytial virus (RSV) is the major cause of lower respiratory tract infection in infants, with about half being infected in their first year of life. Yet only 2 to 3% of infants are hospitalized for RSV infection, suggesting that individual susceptibility contributes to disease severity. Previously, we determined that AKR/J (susceptible) mice developed high lung RSV titers and showed delayed weight recovery, whereas C57BL/6J (resistant) mice demonstrated low lung RSV titers and rapid weight recovery. In addition, we have reported that gene-targeted mice lacking the cystic fibrosis transmembrane conductance regulator (Cftr; ATP-binding cassette subfamily C, member 7) are susceptible to RSV infection. For this report, recombinant backcross and F2 progeny derived from C57BL/6J and AKR/J mice were infected with RSV, their lung titers were measured, and quantitative trait locus (QTL) analysis was performed. A major QTL, designated Rsvs1, was identified on proximal mouse chromosome 6 in both recombinant populations. Microarray analysis comparing lung transcripts of the parental strains during infection identified several candidate genes that mapped to the Rsvs1 interval, including Cftr. These findings add to our understanding of individual RSV susceptibility and strongly support a modifier role for CFTR in RSV infection, a significant cause of respiratory morbidity in infants with cystic fibrosis.
Figures


Similar articles
-
Determinants of host susceptibility to murine respiratory syncytial virus (RSV) disease identify a role for the innate immunity scavenger receptor MARCO gene in human infants.EBioMedicine. 2016 Sep;11:73-84. doi: 10.1016/j.ebiom.2016.08.011. Epub 2016 Aug 6. EBioMedicine. 2016. PMID: 27554839 Free PMC article.
-
Genetic susceptibility to respiratory syncytial virus infection in inbred mice.J Med Virol. 2002 May;67(1):92-100. doi: 10.1002/jmv.2196. J Med Virol. 2002. PMID: 11920822
-
Respiratory syncytial virus infection in a murine model of cystic fibrosis.J Med Virol. 2006 May;78(5):651-8. doi: 10.1002/jmv.20589. J Med Virol. 2006. PMID: 16555274
-
Respiratory syncytial virus persistence: evidence in the mouse model.Pediatr Infect Dis J. 2008 Oct;27(10 Suppl):S60-2. doi: 10.1097/INF.0b013e3181684d52. Pediatr Infect Dis J. 2008. PMID: 18820580 Review.
-
Mouse models of chronic lung infection with Pseudomonas aeruginosa: models for the study of cystic fibrosis.Pediatr Pulmonol. 2000 Nov;30(5):413-24. doi: 10.1002/1099-0496(200011)30:5<413::aid-ppul8>3.0.co;2-9. Pediatr Pulmonol. 2000. PMID: 11064433 Review.
Cited by
-
Determinants of host susceptibility to murine respiratory syncytial virus (RSV) disease identify a role for the innate immunity scavenger receptor MARCO gene in human infants.EBioMedicine. 2016 Sep;11:73-84. doi: 10.1016/j.ebiom.2016.08.011. Epub 2016 Aug 6. EBioMedicine. 2016. PMID: 27554839 Free PMC article.
-
Respiratory syncytial virus--a comprehensive review.Clin Rev Allergy Immunol. 2013 Dec;45(3):331-79. doi: 10.1007/s12016-013-8368-9. Clin Rev Allergy Immunol. 2013. PMID: 23575961 Free PMC article. Review.
-
The transcription factor NRSF contributes to epileptogenesis by selective repression of a subset of target genes.Elife. 2014 Aug 12;3:e01267. doi: 10.7554/eLife.01267. Elife. 2014. PMID: 25117540 Free PMC article.
-
Multigenic control and sex bias in host susceptibility to spore-induced pulmonary anthrax in mice.Infect Immun. 2011 Aug;79(8):3204-15. doi: 10.1128/IAI.01389-10. Epub 2011 May 31. Infect Immun. 2011. PMID: 21628518 Free PMC article.
-
Quantitative genetics in the study of virus-induced disease.Adv Virus Res. 2014;88:193-225. doi: 10.1016/B978-0-12-800098-4.00004-0. Adv Virus Res. 2014. PMID: 24373313 Free PMC article. Review.
References
-
- Abman, S. H., J. W. Ogle, N. Butler-Simon, C. M. Rumack, and F. J. Accurso. 1988. Role of respiratory syncytial virus in early hospitalizations for respiratory distress of young infants with cystic fibrosis. J. Pediatr. 113:826-830. - PubMed
-
- Armstrong, D., K. Grimwood, J. B. Carlin, R. Carzino, J. Hull, A. Olinsky, and P. D. Phelan. 1998. Severe viral respiratory infections in infants with cystic fibrosis. Pediatr. Pulmonol. 26:371-379. - PubMed
-
- Benjamini, Y., and Y. Hochberg. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57:289-300.
-
- Boyce, T. G., B. G. Mellen, E. F. Mitchel, Jr., P. F. Wright, and M. R. Griffin. 2000. Rates of hospitalization for respiratory syncytial virus infection among children in Medicaid. J. Pediatr. 137:865-870. - PubMed
-
- Broman, K. W., H. Wu, S. Sen, and G. A. Churchill. 2003. R/qtl: QTL mapping in experimental crosses. Bioinformatics 19:889-890. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- AI046556/AI/NIAID NIH HHS/United States
- R01 HL085655/HL/NHLBI NIH HHS/United States
- R01 HL075562/HL/NHLBI NIH HHS/United States
- HL085655/HL/NHLBI NIH HHS/United States
- R01 HL077763/HL/NHLBI NIH HHS/United States
- HG003749/HG/NHGRI NIH HHS/United States
- R21 LM009662/LM/NLM NIH HHS/United States
- R01 AI046556/AI/NIAID NIH HHS/United States
- LM009662/LM/NLM NIH HHS/United States
- HL077763/HL/NHLBI NIH HHS/United States
- R56 HL075562/HL/NHLBI NIH HHS/United States
- HL075562/HL/NHLBI NIH HHS/United States
- R01 HG003749/HG/NHGRI NIH HHS/United States
- U01 ES015675/ES/NIEHS NIH HHS/United States
- ES015675/ES/NIEHS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

