Janus-faced enzymes yeast Tgl3p and Tgl5p catalyze lipase and acyltransferase reactions
- PMID: 20016004
- PMCID: PMC2820416
- DOI: 10.1091/mbc.e09-09-0775
Janus-faced enzymes yeast Tgl3p and Tgl5p catalyze lipase and acyltransferase reactions
Abstract
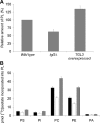
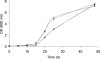
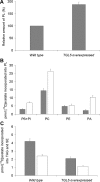
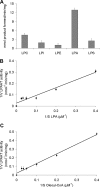
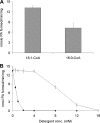
In the yeast, mobilization of triacylglycerols (TAGs) is facilitated by the three TAG lipases Tgl3p, Tgl4p, and Tgl5p. Motif search analysis, however, indicated that Tgl3p and Tgl5p do not only contain the TAG lipase motif GXSXG but also an H-(X)(4)-D acyltransferase motif. Interestingly, lipid analysis revealed that deletion of TGL3 resulted in a decrease and overexpression of TGL3 in an increase of glycerophospholipids. Similar results were obtained with TGL5. Therefore, we tested purified Tgl3p and Tgl5p for acyltransferase activity. Indeed, both enzymes not only exhibited lipase activity but also catalyzed acylation of lysophosphatidylethanolamine and lysophosphatidic acid, respectively. Experiments using variants of Tgl3p created by site-directed mutagenesis clearly demonstrated that the two enzymatic activities act independently of each other. We also showed that Tgl3p is important for efficient sporulation of yeast cells, but rather through its acyltransferase than lipase activity. In summary, our results demonstrate that yeast Tgl3p and Tgl5p play a dual role in lipid metabolism contributing to both anabolic and catabolic processes.
Figures



References
-
- Athenstaedt K., Daum G. YMR313c/TGL3 encodes a novel triacylglycerol lipase located in lipid particles of Saccharomyces cerevisiae. J. Biol. Chem. 2003;278:23317–23323. - PubMed
-
- Athenstaedt K., Daum G. Tgl4p and Tgl5p, two triacylglycerol lipases of the yeast Saccharomyces cerevisiae are localized to lipid particles. J. Biol. Chem. 2005;280:37301–37309. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

