Integrin activation by Fam38A uses a novel mechanism of R-Ras targeting to the endoplasmic reticulum
- PMID: 20016066
- PMCID: PMC2794710
- DOI: 10.1242/jcs.056424
Integrin activation by Fam38A uses a novel mechanism of R-Ras targeting to the endoplasmic reticulum
Abstract
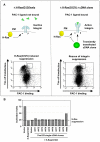
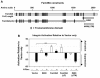
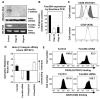
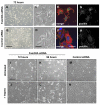
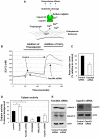
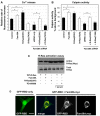
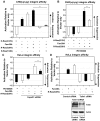
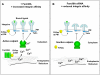
The integrin family of heterodimeric cell-surface receptors are fundamental in cell-cell and cell-matrix adhesion. Changes to either integrin-ligand affinity or integrin gene expression are central to a variety of disease processes, including inflammation, cardiovascular disease and cancer. In screening for novel activators of integrin-ligand affinity we identified the previously uncharacterised multi-transmembrane domain protein Fam38A, located at the endoplasmic reticulum (ER). siRNA knockdown of Fam38A in epithelial cells inactivates endogenous beta1 integrin, reducing cell adhesion. Fam38A mediates integrin activation by recruiting the small GTPase R-Ras to the ER, which activates the calcium-activated protease calpain by increasing Ca(2+) release from cytoplasmic stores. Fam38A-induced integrin activation is blocked by inhibition of either R-Ras or calpain activity, or by siRNA knockdown of talin, a well-described calpain substrate. This highlights a novel mechanism for integrin activation by Fam38A, utilising calpain and R-Ras signalling from the ER. These data represent the first description of a novel spatial regulator of R-Ras, of an alternative integrin activation-suppression pathway based on direct relocalisation of R-Ras to the ER, and of a mechanism linking R-Ras and calpain signalling from the ER with modulation of integrin-ligand affinity.
Figures


Similar articles
-
Loss of the integrin-activating transmembrane protein Fam38A (Piezo1) promotes a switch to a reduced integrin-dependent mode of cell migration.PLoS One. 2012;7(7):e40346. doi: 10.1371/journal.pone.0040346. Epub 2012 Jul 5. PLoS One. 2012. PMID: 22792288 Free PMC article.
-
R-Ras regulates beta1-integrin trafficking via effects on membrane ruffling and endocytosis.BMC Cell Biol. 2010 Feb 18;11:14. doi: 10.1186/1471-2121-11-14. BMC Cell Biol. 2010. PMID: 20167113 Free PMC article.
-
RIAM activates integrins by linking talin to ras GTPase membrane-targeting sequences.J Biol Chem. 2009 Feb 20;284(8):5119-27. doi: 10.1074/jbc.M807117200. Epub 2008 Dec 19. J Biol Chem. 2009. PMID: 19098287 Free PMC article.
-
Mammalian NOTCH-1 activates beta1 integrins via the small GTPase R-Ras.J Biol Chem. 2007 Sep 28;282(39):28991-29001. doi: 10.1074/jbc.M703601200. Epub 2007 Jul 30. J Biol Chem. 2007. PMID: 17664272
-
Differential role of beta(1C) and beta(1A) integrin cytoplasmic variants in modulating focal adhesion kinase, protein kinase B/AKT, and Ras/Mitogen-activated protein kinase pathways.Mol Biol Cell. 2000 Jul;11(7):2235-49. doi: 10.1091/mbc.11.7.2235. Mol Biol Cell. 2000. PMID: 10888665 Free PMC article.
Cited by
-
Mechanical dynamics in live cells and fluorescence-based force/tension sensors.Biochim Biophys Acta. 2015 Aug;1853(8):1889-904. doi: 10.1016/j.bbamcr.2015.05.001. Epub 2015 May 6. Biochim Biophys Acta. 2015. PMID: 25958335 Free PMC article. Review.
-
Integrins in mechanotransduction.Curr Opin Cell Biol. 2013 Oct;25(5):613-8. doi: 10.1016/j.ceb.2013.05.006. Epub 2013 Jun 21. Curr Opin Cell Biol. 2013. PMID: 23797029 Free PMC article. Review.
-
The Mechanosensitive Ion Channel Piezo1 Significantly Mediates In Vitro Ultrasonic Stimulation of Neurons.iScience. 2019 Nov 22;21:448-457. doi: 10.1016/j.isci.2019.10.037. Epub 2019 Oct 23. iScience. 2019. PMID: 31707258 Free PMC article.
-
Piezo1 Channel as a Potential Target for Hindering Cardiac Fibrotic Remodeling.Int J Mol Sci. 2022 Jul 22;23(15):8065. doi: 10.3390/ijms23158065. Int J Mol Sci. 2022. PMID: 35897650 Free PMC article. Review.
-
Endomembranes: Unsung Heroes of Mechanobiology?Front Bioeng Biotechnol. 2020 Oct 22;8:597721. doi: 10.3389/fbioe.2020.597721. eCollection 2020. Front Bioeng Biotechnol. 2020. PMID: 33195167 Free PMC article. Review.
References
-
- Ada-Nguema, A. S., Xenias, H., Hofman, J. M., Wiggins, C. H., Sheetz, M. P. and Keely, P. J. (2006). The small GTPase R-Ras regulates organization of actin and drives membrane protrusions through the activity of PLCepsilon. J. Cell Sci. 119, 1307-1319. - PubMed
-
- Altznauer, F., Conus, S., Cavalli, A., Folkers, G. and Simon, H. U. (2004). Calpain-1 regulates Bax and subsequent Smac-dependent caspase-3 activation in neutrophil apoptosis. J. Biol. Chem. 279, 5947-5957. - PubMed
-
- Bhatt, A., Kaverina, I., Otey, C. and Huttenlocher, A. (2002). Regulation of focal complex composition and disass.e.m.bly by the calcium-dependent protease calpain. J. Cell Sci. 115, 3415-3425. - PubMed
-
- Carragher, N. O., Walker, S. M., Scott Carragher, L. A., Harris, F., Sawyer, T. K., Brunton, V. G., Ozanne, B. W. and Frame, M. C. (2006). Calpain 2 and Src dependence distinguishes mesenchymal and amoeboid modes of tumour cell invasion: a link to integrin function. Oncogene 25, 5726-5740. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

