Identification of sister chromatids by DNA template strand sequences
- PMID: 20016487
- PMCID: PMC3757939
- DOI: 10.1038/nature08644
Identification of sister chromatids by DNA template strand sequences
Abstract
It is generally assumed that sister chromatids are genetically and functionally identical and that segregation to daughter cells is a random process. However, functional differences between sister chromatids regulate daughter cell fate in yeast and sister chromatid segregation is not random in Escherichia coli. Differentiated sister chromatids, coupled with non-random segregation, have been proposed to regulate cell fate during the development of multicellular organisms. This hypothesis has not been tested because molecular features to reliably distinguish between sister chromatids are not obvious. Here we show that parental 'Watson' and 'Crick' DNA template strands can be identified in sister chromatids of murine metaphase chromosomes using CO-FISH (chromosome orientation fluorescence in situ hybridization) with unidirectional probes specific for centromeric and telomeric repeats. All chromosomes were found to have a uniform orientation with the 5' end of the short arm on the same strand as T-rich major satellite repeats. The invariable orientation of repetitive DNA was used to differentially label sister chromatids and directly study mitotic segregation patterns in different cell types. Whereas sister chromatids appeared to be randomly distributed between daughter cells in cultured lung fibroblasts and embryonic stem cells, significant non-random sister chromatid segregation was observed in a subset of colon crypt epithelial cells, including cells outside positions reported for colon stem cells. Our results establish that DNA template sequences can be used to distinguish sister chromatids and follow their mitotic segregation in vivo.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


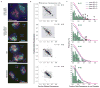
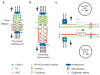
References
-
- Klar AJ. Differentiated parental DNA strands confer developmental asymmetry on daughter cells in fission yeast. Nature. 1987;326 (6112):466–470. - PubMed
-
- White MA, Eykelenboom JK, Lopez-Vernaza MA, Wilson E, Leach DR. Non-random segregation of sister chromosomes in Escherichia coli. Nature. 2008;455 (7217):1248–1250. - PubMed
-
- Lansdorp PM. Immortal strands? Give me a break. Cell. 2007;129 (7):1244–1247. - PubMed
-
- Meyne J, Goodwin EH. Strand-specific fluorescence in situ hybridization for determining orientation and direction of DNA sequences. Methods Mol Biol. 1994;33:141–145. - PubMed
-
- Barker N, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449 (7165):1003–1007. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

