Phosphorylation-mediated control of histone chaperone ASF1 levels by Tousled-like kinases
- PMID: 20016786
- PMCID: PMC2791443
- DOI: 10.1371/journal.pone.0008328
Phosphorylation-mediated control of histone chaperone ASF1 levels by Tousled-like kinases
Abstract
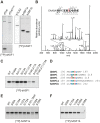
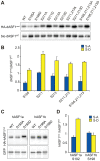
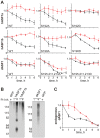
Histone chaperones are at the hub of a diverse interaction networks integrating a plethora of chromatin modifying activities. Histone H3/H4 chaperone ASF1 is a target for cell-cycle regulated Tousled-like kinases (TLKs) and both proteins cooperate during chromatin replication. However, the precise role of post-translational modification of ASF1 remained unclear. Here, we identify the TLK phosphorylation sites for both Drosophila and human ASF1 proteins. Loss of TLK-mediated phosphorylation triggers hASF1a and dASF1 degradation by proteasome-dependent and independent mechanisms respectively. Consistent with this notion, introduction of phosphorylation-mimicking mutants inhibits hASF1a and dASF1 degradation. Human hASF1b is also targeted for proteasome-dependent degradation, but its stability is not affected by phosphorylation indicating that other mechanisms are likely to be involved in control of hASF1b levels. Together, these results suggest that ASF1 cellular levels are tightly controlled by distinct pathways and provide a molecular mechanism for post-translational regulation of dASF1 and hASF1a by TLK kinases.
Conflict of interest statement
Figures

References
-
- De Koning L, Corpet A, Haber JE, Almouzni G. Histone chaperones: an escort network regulating histone traffic. Nat Struct Mol Biol. 2007;14:997–1007. - PubMed
-
- Moshkin YM, Kan TW, Goodfellow H, Bezstarosti K, Maeda RK, et al. Histone chaperones ASF1 and NAP1 differentially modulate removal of active histone marks by LID-RPD3 complexes during NOTCH silencing. Mol Cell. 2009;35:782–793. - PubMed
-
- Tyler JK, Adams CR, Chen SR, Kobayashi R, Kamakaka RT, et al. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature. 1999;402:555–560. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

