Rhodopsin molecular evolution in mammals inhabiting low light environments
- PMID: 20016835
- PMCID: PMC2790605
- DOI: 10.1371/journal.pone.0008326
Rhodopsin molecular evolution in mammals inhabiting low light environments
Abstract
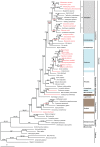
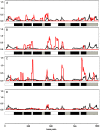
The ecological radiation of mammals to inhabit a variety of light environments is largely attributed to adaptive changes in their visual systems. Visual capabilities are conferred by anatomical features of the eyes as well as the combination and properties of their constituent light sensitive pigments. To test whether evolutionary switches to different niches characterized by dim-light conditions coincided with molecular adaptation of the rod pigment rhodopsin, we sequenced the rhodopsin gene in twenty-two mammals including several bats and subterranean mole-rats. We compared these to thirty-seven published mammal rhodopsin sequences, from species with divergent visual ecologies, including nocturnal, diurnal and aquatic groups. All taxa possessed an intact functional rhodopsin; however, phylogenetic tree reconstruction recovered a gene tree in which rodents were not monophyletic, and also in which echolocating bats formed a monophyletic group. These conflicts with the species tree appear to stem from accelerated evolution in these groups, both of which inhabit low light environments. Selection tests confirmed divergent selection pressures in the clades of subterranean rodents and bats, as well as in marine mammals that live in turbid conditions. We also found evidence of divergent selection pressures among groups of bats with different sensory modalities based on vision and echolocation. Sliding window analyses suggest most changes occur in transmembrane domains, particularly obvious within the pinnipeds; however, we found no obvious pattern between photopic niche and predicted spectral sensitivity based on known critical amino acids. This study indicates that the independent evolution of rhodopsin vision in ecologically specialised groups of mammals has involved molecular evolution at the sequence level, though such changes might not mediate spectral sensitivity directly.
Conflict of interest statement
Figures
Similar articles
-
Divergent positive selection in rhodopsin from lake and riverine cichlid fishes.Mol Biol Evol. 2014 May;31(5):1149-65. doi: 10.1093/molbev/msu064. Epub 2014 Feb 6. Mol Biol Evol. 2014. PMID: 24509690
-
Parallel and convergent evolution of the dim-light vision gene RH1 in bats (Order: Chiroptera).PLoS One. 2010 Jan 21;5(1):e8838. doi: 10.1371/journal.pone.0008838. PLoS One. 2010. PMID: 20098620 Free PMC article.
-
Diversified Mammalian Visuasl Adaptations to Bright- or Dim-Light Environments.Mol Biol Evol. 2023 Apr 4;40(4):msad063. doi: 10.1093/molbev/msad063. Mol Biol Evol. 2023. PMID: 36929909 Free PMC article.
-
Spectral Tuning of Killer Whale (Orcinus orca) Rhodopsin: Evidence for Positive Selection and Functional Adaptation in a Cetacean Visual Pigment.Mol Biol Evol. 2016 Feb;33(2):323-36. doi: 10.1093/molbev/msv217. Epub 2015 Oct 20. Mol Biol Evol. 2016. PMID: 26486871
-
Evolution and spectral tuning of visual pigments in birds and mammals.Philos Trans R Soc Lond B Biol Sci. 2009 Oct 12;364(1531):2941-55. doi: 10.1098/rstb.2009.0044. Philos Trans R Soc Lond B Biol Sci. 2009. PMID: 19720655 Free PMC article. Review.
Cited by
-
Adaptive genomic evolution of opsins reveals that early mammals flourished in nocturnal environments.BMC Genomics. 2018 Feb 5;19(1):121. doi: 10.1186/s12864-017-4417-8. BMC Genomics. 2018. PMID: 29402215 Free PMC article.
-
A system-level, molecular evolutionary analysis of mammalian phototransduction.BMC Evol Biol. 2013 Feb 23;13:52. doi: 10.1186/1471-2148-13-52. BMC Evol Biol. 2013. PMID: 23433342 Free PMC article.
-
Evolution of the Highly Repetitive PEVK Region of Titin Across Mammals.G3 (Bethesda). 2019 Apr 9;9(4):1103-1115. doi: 10.1534/g3.118.200714. G3 (Bethesda). 2019. PMID: 30804022 Free PMC article.
-
Adaptations to a subterranean environment and longevity revealed by the analysis of mole rat genomes.Cell Rep. 2014 Sep 11;8(5):1354-64. doi: 10.1016/j.celrep.2014.07.030. Epub 2014 Aug 28. Cell Rep. 2014. PMID: 25176646 Free PMC article.
-
How to Make a Dolphin: Molecular Signature of Positive Selection in Cetacean Genome.PLoS One. 2013 Jun 19;8(6):e65491. doi: 10.1371/journal.pone.0065491. Print 2013. PLoS One. 2013. PMID: 23840335 Free PMC article.
References
-
- Cott HB. London: Oxford University Press; 1940. Adaptive coloration in animals.
-
- Walls GL. New York: Hafner; 1942. The vertebrate eye and its adaptive radiation.
-
- Wickler W. New York: McGraw-Hill; 1968. Mimicry in plants and animals.
-
- Hinton HE. Natural deception. In: Gregory RL, Gombrich EH, editors. London: Duckworth; 1973.
-
- Ferrari M. Colors for survival: mimicry and camouflage in nature. Charlottesville: Thomasson-Grant 1993
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

