Assessment of genotype imputation methods
- PMID: 20018042
- PMCID: PMC2795949
- DOI: 10.1186/1753-6561-3-s7-s5
Assessment of genotype imputation methods
Abstract
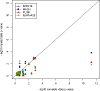
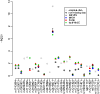
Several methods have been proposed to impute genotypes at untyped markers using observed genotypes and genetic data from a reference panel. We used the Genetic Analysis Workshop 16 rheumatoid arthritis case-control dataset to compare the performance of four of these imputation methods: IMPUTE, MACH, PLINK, and fastPHASE. We compared the methods' imputation error rates and performance of association tests using the imputed data, in the context of imputing completely untyped markers as well as imputing missing genotypes to combine two datasets genotyped at different sets of markers. As expected, all methods performed better for single-nucleotide polymorphisms (SNPs) in high linkage disequilibrium with genotyped SNPs. However, MACH and IMPUTE generated lower imputation error rates than fastPHASE and PLINK. Association tests based on allele "dosage" from MACH and tests based on the posterior probabilities from IMPUTE provided results closest to those based on complete data. However, in both situations, none of the imputation-based tests provide the same level of evidence of association as the complete data at SNPs strongly associated with disease.
Figures

References
-
- Li Y, Abecasis GR. Mach 1.0: rapid haplotype reconstruction and missing genotype inference [abstract 2290/C] Am J Hum Genet. 2006;S79:416.
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

