Use of thermal melt curves to assess the quality of enzyme preparations
- PMID: 20018159
- PMCID: PMC3270315
- DOI: 10.1016/j.ab.2009.12.018
Use of thermal melt curves to assess the quality of enzyme preparations
Abstract
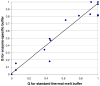
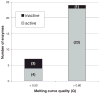
This study sought to determine whether the quality of enzyme preparations can be determined from their melting curves, which may easily be obtained using a fluorescent probe and a standard reverse transcription-polymerase chain reaction (RT-PCR) machine. Thermal melt data on 31 recombinant enzymes from Plasmodium parasites were acquired by incrementally heating them to 90 degrees C and measuring unfolding with a fluorescent dye. Activity assays specific to each enzyme were also performed. Four of the enzymes were denatured to varying degrees with heat and sodium dodecyl sulfate (SDS) prior to the thermal melt and activity assays. In general, melting curve quality was correlated with enzyme activity; enzymes with high-quality curves were found almost uniformly to be active, whereas those with lower quality curves were more varied in their catalytic performance. Inspection of melting curves of bovine xanthine oxidase and Entamoeba histolytica cysteine protease 1 allowed active stocks to be distinguished from inactive stocks, implying that a relationship between melting curve quality and activity persists over a wide range of experimental conditions and species. Our data suggest that melting curves can help to distinguish properly folded proteins from denatured ones and, therefore, may be useful in selecting stocks for further study and in optimizing purification procedures for specific proteins.
Copyright 2009 Elsevier Inc. All rights reserved.
Figures



Similar articles
-
Conformational changes induced by detergents during the refolding of chemically denatured cysteine protease ppEhCP-B9 from Entamoeba histolytica.Biochim Biophys Acta. 2014 Jul;1844(7):1299-306. doi: 10.1016/j.bbapap.2014.04.009. Epub 2014 Apr 24. Biochim Biophys Acta. 2014. PMID: 24769005
-
Molecular and functional characterisation of a stress responsive cysteine protease, EhCP6 from Entamoeba histolytica.Protein Expr Purif. 2015 May;109:55-61. doi: 10.1016/j.pep.2015.02.005. Epub 2015 Feb 10. Protein Expr Purif. 2015. PMID: 25680771
-
Entamoeba histolytica and E. invadens: sulfhydryl-dependent proteolytic activity.J Protozool. 1985 Feb;32(1):163-6. doi: 10.1111/j.1550-7408.1985.tb03032.x. J Protozool. 1985. PMID: 2859367
-
Cysteine proteases: Battling pathogenic parasitic protozoans with omnipresent enzymes.Microbiol Res. 2021 Aug;249:126784. doi: 10.1016/j.micres.2021.126784. Epub 2021 May 6. Microbiol Res. 2021. PMID: 33989978 Review.
-
Real-time multiplex PCR assays.Methods. 2001 Dec;25(4):430-42. doi: 10.1006/meth.2001.1265. Methods. 2001. PMID: 11846612 Review.
Cited by
-
Insights into the slow-onset tight-binding inhibition of Escherichia coli dihydrofolate reductase: detailed mechanistic characterization of pyrrolo [3,2-f] quinazoline-1,3-diamine and its derivatives as novel tight-binding inhibitors.FEBS J. 2015 May;282(10):1922-38. doi: 10.1111/febs.13244. Epub 2015 Mar 6. FEBS J. 2015. PMID: 25703118 Free PMC article.
-
Ligand induced stabilization of the melting temperature of the HSV-1 single-strand DNA binding protein using the thermal shift assay.Biochem Biophys Res Commun. 2014 Nov 28;454(4):604-8. doi: 10.1016/j.bbrc.2014.10.145. Epub 2014 Nov 4. Biochem Biophys Res Commun. 2014. PMID: 25449284 Free PMC article.
-
PoLi: A Virtual Screening Pipeline Based on Template Pocket and Ligand Similarity.J Chem Inf Model. 2015 Aug 24;55(8):1757-70. doi: 10.1021/acs.jcim.5b00232. Epub 2015 Aug 12. J Chem Inf Model. 2015. PMID: 26225536 Free PMC article.
-
Catalytic Properties of Caseinolytic Protease Subunit of Plasmodium knowlesi and Its Inhibition by a Member of δ-Lactone, Hyptolide.Molecules. 2022 Jun 12;27(12):3787. doi: 10.3390/molecules27123787. Molecules. 2022. PMID: 35744912 Free PMC article.
-
Novel small molecule binders of human N-glycanase 1, a key player in the endoplasmic reticulum associated degradation pathway.Bioorg Med Chem. 2016 Oct 1;24(19):4750-4758. doi: 10.1016/j.bmc.2016.08.019. Epub 2016 Aug 13. Bioorg Med Chem. 2016. PMID: 27567076 Free PMC article.
References
-
- Pantoliano MW, Petrella EC, Kwasnoski JD, Lobanov VS, Myslik J, Graf E, Carver T, Asel E, Springer BA, Lane P, Salemme FR. High-density miniaturized thermal shift assays as a general strategy for drug discovery. J Biomol Screen. 2001;6:429–440. - PubMed
-
- Mezzasalma TM, Kranz JK, Chan W, Struble GT, Schalk-Hihi C, Deckman IC, Springer BA, Todd MJ. Enhancing recombinant protein quality and yield by protein stability profiling. J Biomol Screen. 2007;12:418–428. - PubMed
-
- Crowther GJ, Napuli AJ, Thomas AP, Chung DJ, Kovzun KV, Leibly DJ, Castaneda LJ, Bhandari J, Damman CJ, Hui R, Hol WGJ, Buckner FS, Verlinde CLMJ, Zhang Z, Fan E, Van Voorhis WC. Buffer optimization of thermal melt assays of Plasmodium proteins for detection of small-molecule ligands. Journal of Biomolecular Screening. 2009;14 in press. - PMC - PubMed
-
- Bracher A, Eisenreich W, Schramek N, Ritz H, Gotze E, Herrmann A, Gutlich M, Bacher A. Biosynthesis of pteridines. NMR studies on the reaction mechanisms of GTP cyclohydrolase I, pyruvoyltetrahydropterin synthase, and sepiapterin reductase. J Biol Chem. 1998;273:28132–28141. - PubMed
-
- Melendez-Lopez SG, Herdman S, Hirata K, Choi MH, Choe Y, Craik C, Caffrey CR, Hansell E, Chavez-Munguia B, Chen YT, Roush WR, McKerrow J, Eckmann L, Guo J, Stanley SL, Jr, Reed SL. Use of recombinant Entamoeba histolytica cysteine proteinase 1 to identify a potent inhibitor of amebic invasion in a human colonic model. Eukaryot Cell. 2007;6:1130–1136. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

