Gene expression analysis of macrophages that facilitate tumor invasion supports a role for Wnt-signaling in mediating their activity in primary mammary tumors
- PMID: 20018620
- PMCID: PMC3226722
- DOI: 10.4049/jimmunol.0902360
Gene expression analysis of macrophages that facilitate tumor invasion supports a role for Wnt-signaling in mediating their activity in primary mammary tumors
Abstract
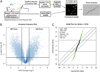
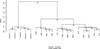
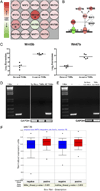
The tumor microenvironment modifies the malignancy of tumors. In solid tumors, this environment is populated by many macrophages that, in genetic studies that depleted these cells from mouse models of breast cancer, were shown to promote tumor progression to malignancy and increase metastatic potential. Mechanistic studies showed that these tumor-promoting effects of macrophages are through the stimulation of tumor cell migration, invasion, intravasation, and enhancement of angiogenesis. Using an in vivo invasion assay, it was demonstrated that invasive carcinoma cells are a unique subpopulation of tumor cells whose invasion and chemotaxis is dependent on the comigration of tumor-associated macrophages (TAMs) with obligate reciprocal signaling through an epidermal growth factor-CSF-1 paracrine loop. In this study, these invasion-promoting macrophages were isolated and subjected to analysis of their transcriptome in comparison with TAMs isolated indiscriminately to function using established macrophage markers. Unsupervised analysis of transcript patterns showed that the invasion-associated TAMs represent a unique subpopulation of TAMs that, by gene ontology criteria, have gene expression patterns related to tissue and organ development. Gene set enrichment analysis showed that these macrophages are also specifically enriched for molecules involved in Wnt-signaling. Previously, it was shown that macrophage-derived Wnt molecules promote vascular remodeling and that tumor cells are highly motile and intravasate around perivascular TAM clusters. Taken together, we conjecture that invasive TAMs link angiogenesis and tumor invasion and that Wnt-signaling plays a role in mediating their activity.
Conflict of interest statement
The authors have no financial conflicts of interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

