Ebolavirus VP35 uses a bimodal strategy to bind dsRNA for innate immune suppression
- PMID: 20018665
- PMCID: PMC2806767
- DOI: 10.1073/pnas.0910547107
Ebolavirus VP35 uses a bimodal strategy to bind dsRNA for innate immune suppression
Abstract
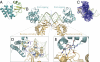
Ebolavirus causes a severe hemorrhagic fever and is divided into five distinct species, of which Reston ebolavirus is uniquely nonpathogenic to humans. Disease caused by ebolavirus is marked by early immunosuppression of innate immune signaling events, involving silencing and sequestration of double-stranded RNA (dsRNA) by the viral protein VP35. Here we present unbound and dsRNA-bound crystal structures of the dsRNA-binding domain of Reston ebolavirus VP35. The structures show that VP35 forms an unusual, asymmetric dimer on dsRNA binding, with each of the monomers binding dsRNA in a different way: one binds the backbone whereas the other caps the terminus. Additional SAXS, DXMS, and dsRNA-binding experiments presented here support a model of cooperative dsRNA recognition in which binding of the first monomer assists binding of the next monomer of the oligomeric VP35 protein. This work illustrates how ebolavirus VP35 could mask key recognition sites of molecules such as RIG-I, MDA-5, and Dicer to silence viral dsRNA in infection.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

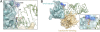


References
-
- Sanchez A, Geisbert TW, Feldmann H. Filoviridae: Marburg and Ebola Viruses. In: Knipe DM, Howley PM, editors. Fields Virology. Philidelphia: Lippincott Williams & Wilkins; 2007. pp. 1409–1448.
-
- Bwaka MA, et al. Ebola hemorrhagic fever in Kikwit, Democratic Republic of the Congo: Clinical observations in 103 patients. J Infect Dis. 1999;179(Suppl 1):S1–S7. - PubMed
-
- Barrette RW, et al. Discovery of swine as a host for the Reston ebolavirus. Science. 2009;325:204–206. - PubMed
-
- CDC. Update: Filovirus infections among persons with occupational exposure to nonhuman primates. MMWR Morb Mortal Wkly. 1990;39:266–7. 273. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
- R21AI053423/AI/NIAID NIH HHS/United States
- R21 CA118595/CA/NCI NIH HHS/United States
- CA099835/CA/NCI NIH HHS/United States
- R21 AI076961/AI/NIAID NIH HHS/United States
- T32 AI007244/AI/NIAID NIH HHS/United States
- 2T32AI007244-26/AI/NIAID NIH HHS/United States
- 5T32AI00760/AI/NIAID NIH HHS/United States
- R01 GM086701/GM/NIGMS NIH HHS/United States
- R33 CA099835/CA/NCI NIH HHS/United States
- R01GM086701/GM/NIGMS NIH HHS/United States
- AI076961/AI/NIAID NIH HHS/United States
- CA118595/CA/NCI NIH HHS/United States
- R21 CA099835/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

