Hybridization kinetics is different inside cells
- PMID: 20018715
- PMCID: PMC2793311
- DOI: 10.1073/pnas.0901313106
Hybridization kinetics is different inside cells
Abstract
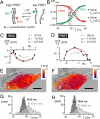
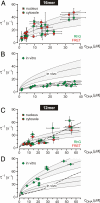
It is generally expected that the kinetics of reactions inside living cells differs from the situation in bulk solutions. Macromolecular crowding and specific binding interactions could change the diffusion properties and the availability of free molecules. Their impact on reaction kinetics in the relevant context of living cells is still elusive, mainly because the difficulty of capturing fast kinetics in vivo. This article shows spatially resolved measurements of DNA hybridization kinetics in single living cells. HeLa cells were transfected with a FRET-labeled dsDNA probe by lipofection. We characterized the hybridization reaction kinetics with a kinetic range of 10 micros to 1 s by a combination of laser-driven temperature oscillations and stroboscopic fluorescence imaging. The time constant of the hybridization depended on DNA concentration within individual cells and between cells. A quantitative analysis of the concentration dependence revealed several-fold accelerated kinetics as compared with free solution for a 16-bp probe and decelerated kinetics for a 12-bp probe. We did not find significant effects of crowding agents on the hybridization kinetics in vitro. Our results suggest that the reaction rates in vivo are specifically modulated by binding interactions for the two probes, possibly triggered by their different lengths. In general, the presented imaging modality of temperature oscillation optical lock-in microscopy allows to probe biomolecular interactions in different cell compartments in living cells for systems biology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



Similar articles
-
Quantitative hybridization kinetics of DNA probes to RNA in solution followed by diffusional fluorescence correlation analysis.Biochemistry. 1996 Aug 6;35(31):10182-93. doi: 10.1021/bi960517g. Biochemistry. 1996. PMID: 8756483
-
Identification of Individual Immobilized DNA Molecules by Their Hybridization Kinetics Using Single-Molecule Fluorescence Imaging.Anal Chem. 2018 Apr 17;90(8):5007-5014. doi: 10.1021/acs.analchem.7b04512. Epub 2018 Mar 30. Anal Chem. 2018. PMID: 29577717
-
Fluorescence Dynamics of a FRET Probe Designed for Crowding Studies.J Phys Chem B. 2017 Jun 15;121(23):5688-5698. doi: 10.1021/acs.jpcb.7b01306. Epub 2017 May 31. J Phys Chem B. 2017. PMID: 28520430
-
Detecting RNA/DNA hybridization using double-labeled donor probes with enhanced fluorescence resonance energy transfer signals.Methods Mol Biol. 2006;335:43-56. doi: 10.1385/1-59745-069-3:43. Methods Mol Biol. 2006. PMID: 16785619 Review.
-
Factors and methods to modulate DNA hybridization kinetics.Biotechnol J. 2021 Nov;16(11):e2000338. doi: 10.1002/biot.202000338. Epub 2021 Sep 2. Biotechnol J. 2021. PMID: 34411451 Review.
Cited by
-
Assembly of Biologically Functional Structures by Nucleic Acid Templating: Implementation of a Strategy to Overcome Inhibition by Template Excess.Molecules. 2022 Oct 12;27(20):6831. doi: 10.3390/molecules27206831. Molecules. 2022. PMID: 36296424 Free PMC article.
-
Lipid vesicles chaperone an encapsulated RNA aptamer.Nat Commun. 2018 Jun 13;9(1):2313. doi: 10.1038/s41467-018-04783-8. Nat Commun. 2018. PMID: 29899431 Free PMC article.
-
Expanding discriminative dimensions for analysis and imaging.Chem Sci. 2015 May 1;6(5):2968-2978. doi: 10.1039/c4sc03955f. Epub 2015 Mar 18. Chem Sci. 2015. PMID: 28706678 Free PMC article.
-
DNA nanotechnology from the test tube to the cell.Nat Nanotechnol. 2015 Sep;10(9):748-60. doi: 10.1038/nnano.2015.195. Nat Nanotechnol. 2015. PMID: 26329111 Review.
-
A physics-based approach of coarse-graining the cytoplasm of Escherichia coli (CGCYTO).Biophys J. 2012 May 16;102(10):2353-61. doi: 10.1016/j.bpj.2012.04.010. Epub 2012 May 15. Biophys J. 2012. PMID: 22677389 Free PMC article.
References
-
- Lippincott-Schwartz J, Snapp E, Kenworthy A. Studying protein dynamics in living cells. Nat Rev Mol Cell Biol. 2001;2:444–456. - PubMed
-
- Miyawaki A. Visualization of the spatial and temporal dynamics of intracellular signaling. Dev Cell. 2003;4:295–305. - PubMed
-
- Giepmans BNG, Adams SR, Ellisman MH, Tsien RY. The fluorescent toolbox for assessing protein location and function. Science. 2006;312:217–224. - PubMed
-
- Ideker T, Galitski T, Hood L. A new approach to decoding life: Systems biology. Annu Rev Genomics Hum Genet. 2001;2:343–372. - PubMed
-
- Di Ventura B, Lemerle C, Michalodimitrakis K, Serrano L. From in vivo to in silico biology and back. Nature. 2006;443:527–533. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

