Complex regulation of the TIR1/AFB family of auxin receptors
- PMID: 20018756
- PMCID: PMC2799741
- DOI: 10.1073/pnas.0911967106
Complex regulation of the TIR1/AFB family of auxin receptors
Abstract
Auxin regulates most aspects of plant growth and development. The hormone is perceived by the TIR1/AFB family of F-box proteins acting in concert with the Aux/IAA transcriptional repressors. Arabidopsis plants that lack members of the TIR1/AFB family are auxin resistant and display a variety of growth defects. However, little is known about the functional differences between individual members of the family. Phylogenetic studies reveal that the TIR1/AFB proteins are conserved across land plant lineages and fall into four clades. Three of these subgroups emerged before separation of angiosperms and gymnosperms whereas the last emerged before the monocot-eudicot split. This evolutionary history suggests that the members of each clade have distinct functions. To explore this possibility in Arabidopsis, we have analyzed a range of mutant genotypes, generated promoter swap transgenic lines, and performed in vitro binding assays between individual TIR1/AFB and Aux/IAA proteins. Our results indicate that the TIR1/AFB proteins have distinct biochemical activities and that TIR1 and AFB2 are the dominant auxin receptors in the seedling root. Further, we demonstrate that TIR1, AFB2, and AFB3, but not AFB1 exhibit significant posttranscriptional regulation. The microRNA miR393 is expressed in a pattern complementary to that of the auxin receptors and appears to regulate TIR1/AFB expression. However our data suggest that this regulation is complex. Our results suggest that differences between members of the auxin receptor family may contribute to the complexity of auxin response.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
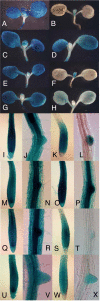
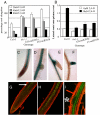
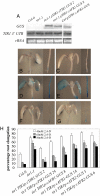

References
-
- Benjamins R, Scheres B. Auxin: The looping star in plant development. Annu Rev Plant Biol. 2008;59:443–465. - PubMed
-
- Dharmasiri N, et al. Plant development is regulated by a family of auxin receptor F box proteins. Dev Cell. 2005;9(1):109–119. - PubMed
-
- Mockaitis K, Estelle M. Auxin receptors and plant development: A new signaling paradigm. Annu Rev Cell Dev Biol. 2008;24:55–80. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

