Crystal structure and computational analyses provide insights into the catalytic mechanism of 2,4-diacetylphloroglucinol hydrolase PhlG from Pseudomonas fluorescens
- PMID: 20018877
- PMCID: PMC2836065
- DOI: 10.1074/jbc.M109.044180
Crystal structure and computational analyses provide insights into the catalytic mechanism of 2,4-diacetylphloroglucinol hydrolase PhlG from Pseudomonas fluorescens
Abstract
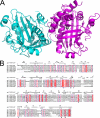
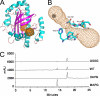
2,4-Diacetylphloroglucinol hydrolase PhlG from Pseudomonas fluorescens catalyzes hydrolytic carbon-carbon (C-C) bond cleavage of the antibiotic 2,4-diacetylphloroglucinol to form monoacetylphloroglucinol, a rare class of reactions in chemistry and biochemistry. To investigate the catalytic mechanism of this enzyme, we determined the three-dimensional structure of PhlG at 2.0 A resolution using x-ray crystallography and MAD methods. The overall structure includes a small N-terminal domain mainly involved in dimerization and a C-terminal domain of Bet v1-like fold, which distinguishes PhlG from the classical alpha/beta-fold hydrolases. A dumbbell-shaped substrate access tunnel was identified to connect a narrow interior amphiphilic pocket to the exterior solvent. The tunnel is likely to undergo a significant conformational change upon substrate binding to the active site. Structural analysis coupled with computational docking studies, site-directed mutagenesis, and enzyme activity analysis revealed that cleavage of the 2,4-diacetylphloroglucinol C-C bond proceeds via nucleophilic attack by a water molecule, which is coordinated by a zinc ion. In addition, residues Tyr(121), Tyr(229), and Asn(132), which are predicted to be hydrogen-bonded to the hydroxyl groups and unhydrolyzed acetyl group, can finely tune and position the bound substrate in a reactive orientation. Taken together, these results revealed the active sites and zinc-dependent hydrolytic mechanism of PhlG and explained its substrate specificity as well.
Figures
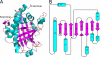

Similar articles
-
Transcriptional Regulator PhlH Modulates 2,4-Diacetylphloroglucinol Biosynthesis in Response to the Biosynthetic Intermediate and End Product.Appl Environ Microbiol. 2017 Oct 17;83(21):e01419-17. doi: 10.1128/AEM.01419-17. Print 2017 Nov 1. Appl Environ Microbiol. 2017. PMID: 28821548 Free PMC article.
-
Characterization of PhlG, a hydrolase that specifically degrades the antifungal compound 2,4-diacetylphloroglucinol in the biocontrol agent Pseudomonas fluorescens CHA0.Appl Environ Microbiol. 2006 Jan;72(1):418-27. doi: 10.1128/AEM.72.1.418-427.2006. Appl Environ Microbiol. 2006. PMID: 16391073 Free PMC article.
-
Molecular and catalytic properties of 2,4-diacetylphloroglucinol hydrolase (PhlG) from Pseudomonas sp. YGJ3.Biosci Biotechnol Biochem. 2012;76(6):1239-41. doi: 10.1271/bbb.120054. Epub 2012 Jun 7. Biosci Biotechnol Biochem. 2012. PMID: 22790955
-
Structure and function analysis of Pseudomonas plant cell wall hydrolases.Prog Nucleic Acid Res Mol Biol. 1998;61:211-41. doi: 10.1016/s0079-6603(08)60828-4. Prog Nucleic Acid Res Mol Biol. 1998. PMID: 9752722 Review.
-
Structure, mechanism, and substrate specificity of kynureninase.Biochim Biophys Acta. 2011 Nov;1814(11):1481-8. doi: 10.1016/j.bbapap.2010.12.003. Epub 2010 Dec 15. Biochim Biophys Acta. 2011. PMID: 21167323 Free PMC article. Review.
Cited by
-
Crystallization and preliminary X-ray diffraction analysis of a putative carbon-carbon bond hydrolase from Mycobacterium abscessus 103.Acta Crystallogr F Struct Biol Commun. 2015 Feb;71(Pt 2):239-42. doi: 10.1107/S2053230X15001612. Epub 2015 Jan 28. Acta Crystallogr F Struct Biol Commun. 2015. PMID: 25664803 Free PMC article.
-
The Chemistry of Gut Microbial Metabolism of Polyphenols.Phytochem Rev. 2016 Jun;15(3):425-444. doi: 10.1007/s11101-016-9459-z. Epub 2016 Mar 11. Phytochem Rev. 2016. PMID: 27274718 Free PMC article.
-
Partial purification and characterization of 2, 4-diacetylphloroglucinol producing Pseudomonas fluorescens VSMKU3054 against bacterial wilt disease of tomato.Saudi J Biol Sci. 2021 Apr;28(4):2155-2167. doi: 10.1016/j.sjbs.2021.02.073. Epub 2021 Mar 4. Saudi J Biol Sci. 2021. PMID: 33911932 Free PMC article.
-
Transcriptional Regulator PhlH Modulates 2,4-Diacetylphloroglucinol Biosynthesis in Response to the Biosynthetic Intermediate and End Product.Appl Environ Microbiol. 2017 Oct 17;83(21):e01419-17. doi: 10.1128/AEM.01419-17. Print 2017 Nov 1. Appl Environ Microbiol. 2017. PMID: 28821548 Free PMC article.
-
Posttranscriptional regulation of 2,4-diacetylphloroglucinol production by GidA and TrmE in Pseudomonas fluorescens 2P24.Appl Environ Microbiol. 2014 Jul;80(13):3972-81. doi: 10.1128/AEM.00455-14. Epub 2014 Apr 18. Appl Environ Microbiol. 2014. PMID: 24747907 Free PMC article.
References
-
- Haas D., Keel C. (2003) Annu. Rev. Phytopathol. 41, 117–153 - PubMed
-
- Haas D., Défago G. (2005) Nat. Rev. Microbiol. 3, 307–319 - PubMed
-
- Broadbent D., Mabelis R. P., Spencer H. (1976) Phytochemistry 15, 1785
-
- Keel C., Wirthner P., Oberhänsli T., Voisard C., Burger U., Haas D., Défago G. (1990) Symbiosis 9, 327–341
-
- Keel C., Schnider U., Maurhofer M., Voisard C., Laville J., Burger U., Wirthner P., Haas D., Défago G. (1992) Mol. Plant-Microbe Interact. 5, 4–13
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

