Processing of the matricellular protein hevin in mouse brain is dependent on ADAMTS4
- PMID: 20018883
- PMCID: PMC2820812
- DOI: 10.1074/jbc.M109.070318
Processing of the matricellular protein hevin in mouse brain is dependent on ADAMTS4
Abstract
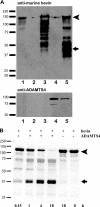
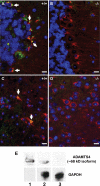
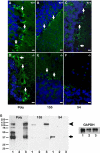
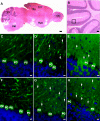
The matricellular SPARC family member hevin (SPARC-like 1/SPARCL-1/SC1/Mast9) contributes to neural development and alters tumor progression in a range of mammalian models. The distribution of hevin in mouse tissues was reexamined with a novel monoclonal antibody that discriminates between hevin and its ortholog SPARC. We now report proteolysis of hevin in many tissues, with the most extensive processing in the brain. We demonstrate a cleavage site within the hevin sequence for the neural tissue proteinase ADAMTS4. Digestion of hevin by ADAMTS4 in vitro produced fragments similar to those present in brain lysates. Monoclonal antibodies revealed a SPARC-like fragment generated from hevin that was co-localized with ADAMTS4 in vivo. We show that proteolysis of hevin by ADAMTS4 in the mouse cerebellum is important for the normal development of this tissue. In conclusion, we have identified the fragmentation of hevin by ADAMTS4 in the mouse brain and propose that this specific proteolysis is integral to cell morphology and extracellular matrix deposition in the developing brain.
Figures


Similar articles
-
Proteolysis of the matricellular protein hevin by matrix metalloproteinase-3 produces a SPARC-like fragment (SLF) associated with neovasculature in a murine glioma model.J Cell Biochem. 2011 Nov;112(11):3093-102. doi: 10.1002/jcb.23235. J Cell Biochem. 2011. PMID: 21688302 Free PMC article.
-
Cartography of hevin-expressing cells in the adult brain reveals prominent expression in astrocytes and parvalbumin neurons.Brain Struct Funct. 2019 Apr;224(3):1219-1244. doi: 10.1007/s00429-019-01831-x. Epub 2019 Jan 17. Brain Struct Funct. 2019. PMID: 30656447
-
Expression and characterization of murine hevin (SC1), a member of the SPARC family of matricellular proteins.J Histochem Cytochem. 2004 Jun;52(6):735-48. doi: 10.1369/jhc.3A6245.2004. J Histochem Cytochem. 2004. PMID: 15150282
-
Hevin/SC1, a matricellular glycoprotein and potential tumor-suppressor of the SPARC/BM-40/Osteonectin family.Int J Biochem Cell Biol. 2004 Jun;36(6):991-6. doi: 10.1016/j.biocel.2004.01.017. Int J Biochem Cell Biol. 2004. PMID: 15094114 Review.
-
The procollagen N-proteinases ADAMTS2, 3 and 14 in pathophysiology.Matrix Biol. 2015 May-Jul;44-46:46-53. doi: 10.1016/j.matbio.2015.04.001. Epub 2015 Apr 8. Matrix Biol. 2015. PMID: 25863161 Review.
Cited by
-
The Matricellular Protein Hevin Is Involved in Alcohol Use Disorder.Biomolecules. 2023 Jan 25;13(2):234. doi: 10.3390/biom13020234. Biomolecules. 2023. PMID: 36830603 Free PMC article.
-
Control of excitatory CNS synaptogenesis by astrocyte-secreted proteins Hevin and SPARC.Proc Natl Acad Sci U S A. 2011 Aug 9;108(32):E440-9. doi: 10.1073/pnas.1104977108. Epub 2011 Jul 25. Proc Natl Acad Sci U S A. 2011. PMID: 21788491 Free PMC article.
-
Species-, organ- and cell-type-dependent expression of SPARCL1 in human and mouse tissues.PLoS One. 2020 May 21;15(5):e0233422. doi: 10.1371/journal.pone.0233422. eCollection 2020. PLoS One. 2020. PMID: 32437418 Free PMC article.
-
Cell-specific and developmental expression of lectican-cleaving proteases in mouse hippocampus and neocortex.J Comp Neurol. 2015 Mar 1;523(4):629-48. doi: 10.1002/cne.23701. Epub 2014 Nov 21. J Comp Neurol. 2015. PMID: 25349050 Free PMC article.
-
Astrocytes Assemble Thalamocortical Synapses by Bridging NRX1α and NL1 via Hevin.Cell. 2016 Jan 14;164(1-2):183-196. doi: 10.1016/j.cell.2015.11.034. Cell. 2016. PMID: 26771491 Free PMC article.
References
-
- Bornstein P., Sage E. H. (2002) Curr. Opin. Cell Biol. 14, 608–616 - PubMed
-
- Brekken R. A., Sage E. H. (2001) Matrix Biol. 19, 816–827 - PubMed
-
- Girard J. P., Springer T. A. (1995) Immunity 2, 113–123 - PubMed
-
- Lively S., Ringuette M. J., Brown I. R. (2007) Neurochem. Res. 32, 65–71 - PubMed
-
- Lively S., Brown I. R. (2008) Neurochem. Res. 33, 1692–1700 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

