An experimentally based computer search identifies unstructured membrane-binding sites in proteins: application to class I myosins, PAKS, and CARMIL
- PMID: 20018884
- PMCID: PMC2820801
- DOI: 10.1074/jbc.M109.066910
An experimentally based computer search identifies unstructured membrane-binding sites in proteins: application to class I myosins, PAKS, and CARMIL
Abstract
Programs exist for searching protein sequences for potential membrane-penetrating segments (hydrophobic regions) and for lipid-binding sites with highly defined tertiary structures, such as PH, FERM, C2, ENTH, and other domains. However, a rapidly growing number of membrane-associated proteins (including cytoskeletal proteins, kinases, GTP-binding proteins, and their effectors) bind lipids through less structured regions. Here, we describe the development and testing of a simple computer search program that identifies unstructured potential membrane-binding sites. Initially, we found that both basic and hydrophobic amino acids, irrespective of sequence, contribute to the binding to acidic phospholipid vesicles of synthetic peptides that correspond to the putative membrane-binding domains of Acanthamoeba class I myosins. Based on these results, we modified a hydrophobicity scale giving Arg- and Lys-positive, rather than negative, values. Using this basic and hydrophobic scale with a standard search algorithm, we successfully identified previously determined unstructured membrane-binding sites in all 16 proteins tested. Importantly, basic and hydrophobic searches identified previously unknown potential membrane-binding sites in class I myosins, PAKs and CARMIL (capping protein, Arp2/3, myosin I linker; a membrane-associated cytoskeletal scaffold protein), and synthetic peptides and protein domains containing these newly identified sites bound to acidic phospholipids in vitro.
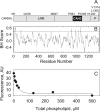
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

