Nucleocytoplasmic shuttling of p62/SQSTM1 and its role in recruitment of nuclear polyubiquitinated proteins to promyelocytic leukemia bodies
- PMID: 20018885
- PMCID: PMC2820819
- DOI: 10.1074/jbc.M109.039925
Nucleocytoplasmic shuttling of p62/SQSTM1 and its role in recruitment of nuclear polyubiquitinated proteins to promyelocytic leukemia bodies
Abstract
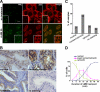
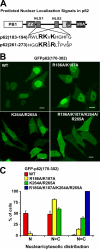
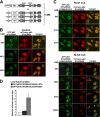
p62, also known as sequestosome1 (SQSTM1), A170, or ZIP, is a multifunctional protein implicated in several signal transduction pathways. p62 is induced by various forms of cellular stress, is degraded by autophagy, and acts as a cargo receptor for autophagic degradation of ubiquitinated targets. It is also suggested to shuttle ubiquitinated proteins for proteasomal degradation. p62 is commonly found in cytosolic protein inclusions in patients with protein aggregopathies, it is up-regulated in several forms of human tumors, and mutations in the gene are linked to classical adult onset Paget disease of the bone. To this end, p62 has generally been considered to be a cytosolic protein, and little attention has been paid to possible nuclear roles of this protein. Here, we present evidence that p62 shuttles continuously between nuclear and cytosolic compartments at a high rate. The protein is also found in nuclear promyelocytic leukemia bodies. We show that p62 contains two nuclear localization signals and a nuclear export signal. Our data suggest that the nucleocytoplasmic shuttling of p62 is modulated by phosphorylations at or near the most important nuclear localization signal, NLS2. The aggregation of p62 in cytosolic bodies also regulates the transport of p62 between the compartments. We found p62 to be essential for accumulation of polyubiquitinated proteins in promyelocytic leukemia bodies upon inhibition of nuclear protein export. Furthermore, p62 contributed to the assembly of proteasome-containing degradative compartments in the vicinity of nuclear aggregates containing polyglutamine-expanded Ataxin1Q84 and to the degradation of Ataxin1Q84.
Figures





Similar articles
-
The Oncogenic Fusion Proteins SET-Nup214 and Sequestosome-1 (SQSTM1)-Nup214 Form Dynamic Nuclear Bodies and Differentially Affect Nuclear Protein and Poly(A)+ RNA Export.J Biol Chem. 2016 Oct 28;291(44):23068-23083. doi: 10.1074/jbc.M116.735340. Epub 2016 Sep 9. J Biol Chem. 2016. PMID: 27613868 Free PMC article.
-
Serine 403 phosphorylation of p62/SQSTM1 regulates selective autophagic clearance of ubiquitinated proteins.Mol Cell. 2011 Oct 21;44(2):279-89. doi: 10.1016/j.molcel.2011.07.039. Mol Cell. 2011. PMID: 22017874
-
p62/sequestosome 1 regulates aggresome formation of pathogenic ataxin-3 with expanded polyglutamine.Int J Mol Sci. 2014 Aug 25;15(9):14997-5010. doi: 10.3390/ijms150914997. Int J Mol Sci. 2014. PMID: 25158237 Free PMC article.
-
Interaction domains of p62: a bridge between p62 and selective autophagy.DNA Cell Biol. 2013 May;32(5):220-7. doi: 10.1089/dna.2012.1915. Epub 2013 Mar 26. DNA Cell Biol. 2013. PMID: 23530606 Review.
-
Autophagic Regulation of p62 is Critical for Cancer Therapy.Int J Mol Sci. 2018 May 8;19(5):1405. doi: 10.3390/ijms19051405. Int J Mol Sci. 2018. PMID: 29738493 Free PMC article. Review.
Cited by
-
p62/SQSTM1 by Binding to Vitamin D Receptor Inhibits Hepatic Stellate Cell Activity, Fibrosis, and Liver Cancer.Cancer Cell. 2016 Oct 10;30(4):595-609. doi: 10.1016/j.ccell.2016.09.004. Cancer Cell. 2016. PMID: 27728806 Free PMC article.
-
The Oncogenic Fusion Proteins SET-Nup214 and Sequestosome-1 (SQSTM1)-Nup214 Form Dynamic Nuclear Bodies and Differentially Affect Nuclear Protein and Poly(A)+ RNA Export.J Biol Chem. 2016 Oct 28;291(44):23068-23083. doi: 10.1074/jbc.M116.735340. Epub 2016 Sep 9. J Biol Chem. 2016. PMID: 27613868 Free PMC article.
-
RTL8 promotes nuclear localization of UBQLN2 to subnuclear compartments associated with protein quality control.Cell Mol Life Sci. 2022 Mar 5;79(3):176. doi: 10.1007/s00018-022-04170-z. Cell Mol Life Sci. 2022. PMID: 35247097 Free PMC article.
-
p62 Is a Potential Biomarker for Risk of Malignant Transformation of Oral Potentially Malignant Disorders (OPMDs).Curr Issues Mol Biol. 2023 Sep 19;45(9):7630-7641. doi: 10.3390/cimb45090480. Curr Issues Mol Biol. 2023. PMID: 37754264 Free PMC article.
-
Intranuclear inclusions in hepatocellular carcinoma contain autophagy-associated proteins and correlate with prolonged survival.J Pathol Clin Res. 2019 Jul;5(3):164-176. doi: 10.1002/cjp2.129. Epub 2019 Apr 8. J Pathol Clin Res. 2019. PMID: 30859721 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

