Characterization of Streptococcus pyogenes beta-NAD+ glycohydrolase: re-evaluation of enzymatic properties associated with pathogenesis
- PMID: 20018886
- PMCID: PMC2820796
- DOI: 10.1074/jbc.M109.070300
Characterization of Streptococcus pyogenes beta-NAD+ glycohydrolase: re-evaluation of enzymatic properties associated with pathogenesis
Abstract
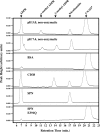
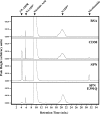
The gram-positive pathogen Streptococcus pyogenes injects a beta-NAD(+) glycohydrolase (SPN) into the cytosol of an infected host cell using the cytolysin-mediated translocation pathway. In this compartment, SPN accelerates the death of the host cell by an unknown mechanism that may involve its beta-NAD(+)-dependent enzyme activities. SPN has been reported to possess the unique characteristic of not only catalyzing hydrolysis of beta-NAD(+), but also carrying out ADP-ribosyl cyclase and ADP-ribosyltransferase activities, making SPN the only beta-NAD(+) glycohydrolase that can catalyze all of these reactions. With the long term goal of understanding how these activities may contribute to pathogenesis, we have further characterized the enzymatic activity of SPN using highly purified recombinant protein. Kinetic studies of the multiple activities of SPN revealed that SPN possessed only beta-NAD(+) hydrolytic activity and lacked detectable ADP-ribosyl cyclase and ADP-ribosyltransferase activities. Similarly, SPN was unable to catalyze cyclic ADPR hydrolysis, and could not catalyze methanolysis or transglycosidation. Kinetic analysis of product inhibition by recombinant SPN demonstrated an ordered uni-bi mechanism, with ADP-ribose being released as a second product. SPN was unaffected by product inhibition using nicotinamide, suggesting that this moiety contributes little to the binding energy of the substrate. Upon transformation, SPN was toxic to Saccharomyces cerevisiae, whereas a glycohydrolase-inactive SPN allowed for viability. Taken together, these data suggest that SPN functions exclusively as a strict beta-NAD(+) glycohydrolase during pathogenesis.
Figures


Similar articles
-
Analysis of polymorphic residues reveals distinct enzymatic and cytotoxic activities of the Streptococcus pyogenes NAD+ glycohydrolase.J Biol Chem. 2013 Jul 5;288(27):20064-75. doi: 10.1074/jbc.M113.481556. Epub 2013 May 20. J Biol Chem. 2013. PMID: 23689507 Free PMC article.
-
High-resolution crystal structure of Streptococcus pyogenes β-NAD⁺ glycohydrolase in complex with its endogenous inhibitor IFS reveals a highly water-rich interface.J Synchrotron Radiat. 2013 Nov;20(Pt 6):962-7. doi: 10.1107/S0909049513020803. Epub 2013 Sep 29. J Synchrotron Radiat. 2013. PMID: 24121349 Free PMC article.
-
Specificity of Streptococcus pyogenes NAD(+) glycohydrolase in cytolysin-mediated translocation.Mol Microbiol. 2006 Nov;62(4):1203-14. doi: 10.1111/j.1365-2958.2006.05430.x. Epub 2006 Oct 17. Mol Microbiol. 2006. PMID: 17042787
-
A novel endogenous inhibitor of the secreted streptococcal NAD-glycohydrolase.PLoS Pathog. 2005 Dec;1(4):e35. doi: 10.1371/journal.ppat.0010035. Epub 2005 Dec 2. PLoS Pathog. 2005. PMID: 16333395 Free PMC article. Review.
-
SIR2: the biochemical mechanism of NAD(+)-dependent protein deacetylation and ADP-ribosyl enzyme intermediates.Curr Med Chem. 2004 Apr;11(7):807-26. doi: 10.2174/0929867043455675. Curr Med Chem. 2004. PMID: 15078167 Review.
Cited by
-
The tuberculosis necrotizing toxin is an NAD+ and NADP+ glycohydrolase with distinct enzymatic properties.J Biol Chem. 2019 Mar 1;294(9):3024-3036. doi: 10.1074/jbc.RA118.005832. Epub 2018 Dec 28. J Biol Chem. 2019. PMID: 30593509 Free PMC article.
-
Structural basis underlying the synergism of NADase and SLO during group A Streptococcus infection.Commun Biol. 2023 Jan 31;6(1):124. doi: 10.1038/s42003-023-04502-0. Commun Biol. 2023. PMID: 36721030 Free PMC article.
-
A novel cholesterol-insensitive mode of membrane binding promotes cytolysin-mediated translocation by Streptolysin O.Mol Microbiol. 2014 Nov;94(3):675-87. doi: 10.1111/mmi.12786. Epub 2014 Sep 23. Mol Microbiol. 2014. PMID: 25196983 Free PMC article.
-
Binding of NAD+-Glycohydrolase to Streptolysin O Stabilizes Both Toxins and Promotes Virulence of Group A Streptococcus.mBio. 2017 Sep 12;8(5):e01382-17. doi: 10.1128/mBio.01382-17. mBio. 2017. PMID: 28900022 Free PMC article.
-
Dissemination of pathogenic bacteria is reinforced by a MARTX toxin effector duet.Nat Commun. 2024 Jul 23;15(1):6218. doi: 10.1038/s41467-024-50650-0. Nat Commun. 2024. PMID: 39043696 Free PMC article.
References
-
- Lin H. (2007) Org. Biomol. Chem. 5, 2541–2554 - PubMed
-
- Ying W. (2008) Antioxid. Redox Signal. 10, 179–206 - PubMed
-
- Schuber F., Lund F. E. (2004) Curr. Mol. Med. 4, 249–261 - PubMed
-
- Lee H. C., Munshi C., Graeff R. (1999) Mol. Cell Biochem. 193, 89–98 - PubMed
-
- Holbourn K. P., Shone C. C., Acharya K. R. (2006) FEBS J. 273, 4579–4593 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

