Phosphorylation of Exo1 modulates homologous recombination repair of DNA double-strand breaks
- PMID: 20019063
- PMCID: PMC2847229
- DOI: 10.1093/nar/gkp1164
Phosphorylation of Exo1 modulates homologous recombination repair of DNA double-strand breaks
Abstract
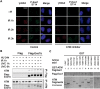
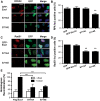
DNA double-strand break (DSB) repair via the homologous recombination pathway is a multi-stage process, which results in repair of the DSB without loss of genetic information or fidelity. One essential step in this process is the generation of extended single-stranded DNA (ssDNA) regions at the break site. This ssDNA serves to induce cell cycle checkpoints and is required for Rad51 mediated strand invasion of the sister chromatid. Here, we show that human Exonuclease 1 (Exo1) is required for the normal repair of DSBs by HR. Cells depleted of Exo1 show chromosomal instability and hypersensitivity to ionising radiation (IR) exposure. We find that Exo1 accumulates rapidly at DSBs and is required for the recruitment of RPA and Rad51 to sites of DSBs, suggesting a role for Exo1 in ssDNA generation. Interestingly, the phosphorylation of Exo1 by ATM appears to regulate the activity of Exo1 following resection, allowing optimal Rad51 loading and the completion of HR repair. These data establish a role for Exo1 in resection of DSBs in human cells, highlighting the critical requirement of Exo1 for DSB repair via HR and thus the maintenance of genomic stability.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

