DNA dynamics play a role as a basal transcription factor in the positioning and regulation of gene transcription initiation
- PMID: 20019064
- PMCID: PMC2847213
- DOI: 10.1093/nar/gkp1084
DNA dynamics play a role as a basal transcription factor in the positioning and regulation of gene transcription initiation
Abstract
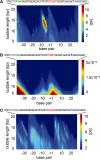
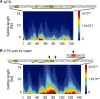
We assess the role of DNA breathing dynamics as a determinant of promoter strength and transcription start site (TSS) location. We compare DNA Langevin dynamic profiles of representative gene promoters, calculated with the extended non-linear PBD model of DNA with experimental data on transcription factor binding and transcriptional activity. Our results demonstrate that DNA dynamic activity at the TSS can be suppressed by mutations that do not affect basal transcription factor binding-DNA contacts. We use this effect to establish the separate contributions of transcription factor binding and DNA dynamics to transcriptional activity. Our results argue against a purely 'transcription factor-centric' view of transcription initiation, suggesting that both DNA dynamics and transcription factor binding are necessary conditions for transcription initiation.
Figures

Similar articles
-
Initiation of transcription at the human terminal deoxynucleotidyl transferase gene promoter: a novel role for the TATA binding protein.Biochemistry. 1994 Sep 13;33(36):11025-32. doi: 10.1021/bi00202a023. Biochemistry. 1994. PMID: 8086419
-
A transcription factor affinity-based code for mammalian transcription initiation.Genome Res. 2009 Apr;19(4):644-56. doi: 10.1101/gr.085449.108. Epub 2009 Jan 13. Genome Res. 2009. PMID: 19141595 Free PMC article.
-
Influence of Rotational Nucleosome Positioning on Transcription Start Site Selection in Animal Promoters.PLoS Comput Biol. 2016 Oct 7;12(10):e1005144. doi: 10.1371/journal.pcbi.1005144. eCollection 2016 Oct. PLoS Comput Biol. 2016. PMID: 27716823 Free PMC article.
-
Role of DNA sequence based structural features of promoters in transcription initiation and gene expression.Curr Opin Struct Biol. 2014 Apr;25:77-85. doi: 10.1016/j.sbi.2014.01.007. Epub 2014 Feb 4. Curr Opin Struct Biol. 2014. PMID: 24503515 Review.
-
What do expression dynamics tell us about the mechanism of transcription?Curr Opin Genet Dev. 2011 Oct;21(5):591-9. doi: 10.1016/j.gde.2011.07.010. Epub 2011 Aug 19. Curr Opin Genet Dev. 2011. PMID: 21862317 Free PMC article. Review.
Cited by
-
Examining DNA breathing with pyDNA-EPBD.Bioinformatics. 2023 Nov 1;39(11):btad699. doi: 10.1093/bioinformatics/btad699. Bioinformatics. 2023. PMID: 37991847 Free PMC article.
-
Nonlinear physics opens a new paradigm for accurate transcription start site prediction.BMC Bioinformatics. 2022 Dec 30;23(1):565. doi: 10.1186/s12859-022-05129-4. BMC Bioinformatics. 2022. PMID: 36585618 Free PMC article.
-
DNA Supercoiling: an Ancestral Regulator of Gene Expression in Pathogenic Bacteria?Comput Struct Biotechnol J. 2019 Jul 26;17:1047-1055. doi: 10.1016/j.csbj.2019.07.013. eCollection 2019. Comput Struct Biotechnol J. 2019. PMID: 31452857 Free PMC article. Review.
-
Non-thermal effects of terahertz radiation on gene expression in mouse stem cells.Biomed Opt Express. 2011 Sep 1;2(9):2679-89. doi: 10.1364/BOE.2.002679. Epub 2011 Aug 23. Biomed Opt Express. 2011. PMID: 21991556 Free PMC article.
-
Analysis of the leakage of gene repression by an artificial TetR-regulated promoter in cyanobacteria.BMC Res Notes. 2015 Sep 19;8:459. doi: 10.1186/s13104-015-1425-0. BMC Res Notes. 2015. PMID: 26387086 Free PMC article.
References
-
- Cheetham GM, Jeruzalmi D, Steitz TA. Structural basis for initiation of transcription from an RNA polymerase-promoter complex. Nature. 1999;399:80–83. - PubMed
-
- Fiedler U, Marc Timmers HT. Peeling by binding or twisting by cranking: models for promoter opening and transcription initiation by RNA polymerase II. Bioessays. 2000;22:316–326. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

