Crystal structure of the eukaryotic strong inward-rectifier K+ channel Kir2.2 at 3.1 A resolution
- PMID: 20019282
- PMCID: PMC2819303
- DOI: 10.1126/science.1180310
Crystal structure of the eukaryotic strong inward-rectifier K+ channel Kir2.2 at 3.1 A resolution
Abstract
Inward-rectifier potassium (K+) channels conduct K+ ions most efficiently in one direction, into the cell. Kir2 channels control the resting membrane voltage in many electrically excitable cells, and heritable mutations cause periodic paralysis and cardiac arrhythmia. We present the crystal structure of Kir2.2 from chicken, which, excluding the unstructured amino and carboxyl termini, is 90% identical to human Kir2.2. Crystals containing rubidium (Rb+), strontium (Sr2+), and europium (Eu3+) reveal binding sites along the ion conduction pathway that are both conductive and inhibitory. The sites correlate with extensive electrophysiological data and provide a structural basis for understanding rectification. The channel's extracellular surface, with large structured turrets and an unusual selectivity filter entryway, might explain the relative insensitivity of eukaryotic inward rectifiers to toxins. These same surface features also suggest a possible approach to the development of inhibitory agents specific to each member of the inward-rectifier K+ channel family.
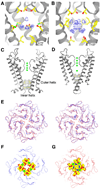
Figures




References
-
- Hodgkin AL, Huxley AF, Katz B. Arch. Sci. Physiol. (Paris) 1949;3:129.
-
- Katz B. Arch. Sci. Physiol. (Paris) 1949;3:285.
-
- Hille B. Ion Channels of Excitable Membranes. Sunderland, MA: Sinauer Associates; 2001.
-
- Fakler B, et al. Cell. 1995 Jan 13;80:149. - PubMed
-
- Lopatin AN, Makhina EN, Nichols CG. Nature. 1994 Nov 24;372:366. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

