Acetylation of Dna2 endonuclease/helicase and flap endonuclease 1 by p300 promotes DNA stability by creating long flap intermediates
- PMID: 20019387
- PMCID: PMC2836044
- DOI: 10.1074/jbc.M109.086397
Acetylation of Dna2 endonuclease/helicase and flap endonuclease 1 by p300 promotes DNA stability by creating long flap intermediates
Abstract
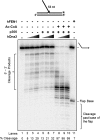
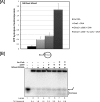
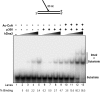
Flap endonuclease 1 (FEN1) and Dna2 endonuclease/helicase (Dna2) sequentially coordinate their nuclease activities for efficient resolution of flap structures that are created during the maturation of Okazaki fragments and repair of DNA damage. Acetylation of FEN1 by p300 inhibits its endonuclease activity, impairing flap cleavage, a seemingly undesirable effect. We now show that p300 also acetylates Dna2, stimulating its 5'-3' endonuclease, the 5'-3' helicase, and DNA-dependent ATPase activities. Furthermore, acetylated Dna2 binds its DNA substrates with higher affinity. Differential regulation of the activities of the two endonucleases by p300 indicates a mechanism in which the acetylase promotes formation of longer flaps in the cell at the same time as ensuring correct processing. Intentional formation of longer flaps mediated by p300 in an active chromatin environment would increase the resynthesis patch size, providing increased opportunity for incorrect nucleotide removal during DNA replication and damaged nucleotide removal during DNA repair. For example, altering the ratio between short and long flap Okazaki fragment processing would be a mechanism for better correction of the error-prone synthesis catalyzed by DNA polymerase alpha.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

