RNA processing and its regulation: global insights into biological networks
- PMID: 20019688
- PMCID: PMC3229837
- DOI: 10.1038/nrg2673
RNA processing and its regulation: global insights into biological networks
Abstract
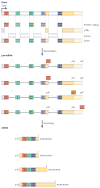
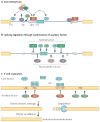
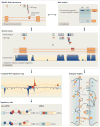
In recent years views of eukaryotic gene expression have been transformed by the finding that enormous diversity can be generated at the RNA level. Advances in technologies for characterizing RNA populations are revealing increasingly complete descriptions of RNA regulation and complexity; for example, through alternative splicing, alternative polyadenylation and RNA editing. New biochemical strategies to map protein-RNA interactions in vivo are yielding transcriptome-wide insights into mechanisms of RNA processing. These advances, combined with bioinformatics and genetic validation, are leading to the generation of functional RNA maps that reveal the rules underlying RNA regulation and networks of biologically coherent transcripts. Together these are providing new insights into molecular cell biology and disease.
Figures
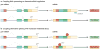
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

