Predicting functional alternative splicing by measuring RNA selection pressure from multigenome alignments
- PMID: 20019791
- PMCID: PMC2784930
- DOI: 10.1371/journal.pcbi.1000608
Predicting functional alternative splicing by measuring RNA selection pressure from multigenome alignments
Abstract
High-throughput methods such as EST sequencing, microarrays and deep sequencing have identified large numbers of alternative splicing (AS) events, but studies have shown that only a subset of these may be functional. Here we report a sensitive bioinformatics approach that identifies exons with evidence of a strong RNA selection pressure ratio (RSPR)--i.e., evolutionary selection against mutations that change only the mRNA sequence while leaving the protein sequence unchanged--measured across an entire evolutionary family, which greatly amplifies its predictive power. Using the UCSC 28 vertebrate genome alignment, this approach correctly predicted half to three-quarters of AS exons that are known binding targets of the NOVA splicing regulatory factor, and predicted 345 strongly selected alternative splicing events in human, and 262 in mouse. These predictions were strongly validated by several experimental criteria of functional AS such as independent detection of the same AS event in other species, reading frame-preservation, and experimental evidence of tissue-specific regulation: 75% (15/20) of a sample of high-RSPR exons displayed tissue specific regulation in a panel of ten tissues, vs. only 20% (4/20) among a sample of low-RSPR exons. These data suggest that RSPR can identify exons with functionally important splicing regulation, and provides biologists with a dataset of over 600 such exons. We present several case studies, including both well-studied examples (GRIN1) and novel examples (EXOC7). These data also show that RSPR strongly outperforms other approaches such as standard sequence conservation (which fails to distinguish amino acid selection pressure from RNA selection pressure), or pairwise genome comparison (which lacks adequate statistical power for predicting individual exons).
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



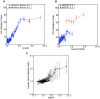

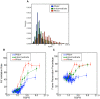

Similar articles
-
Bioinformatics detection of alternative splicing.Methods Mol Biol. 2008;452:179-97. doi: 10.1007/978-1-60327-159-2_9. Methods Mol Biol. 2008. PMID: 18566765 Review.
-
Identification and evolutionary analysis of novel exons and alternative splicing events using cross-species EST-to-genome comparisons in human, mouse and rat.BMC Bioinformatics. 2006 Mar 15;7:136. doi: 10.1186/1471-2105-7-136. BMC Bioinformatics. 2006. PMID: 16536879 Free PMC article.
-
Gene structure prediction and alternative splicing analysis using genomically aligned ESTs.Genome Res. 2001 May;11(5):889-900. doi: 10.1101/gr.155001. Genome Res. 2001. PMID: 11337482 Free PMC article.
-
Evidence of functional selection pressure for alternative splicing events that accelerate evolution of protein subsequences.Proc Natl Acad Sci U S A. 2005 Sep 20;102(38):13526-31. doi: 10.1073/pnas.0501213102. Epub 2005 Sep 12. Proc Natl Acad Sci U S A. 2005. PMID: 16157889 Free PMC article.
-
How prevalent is functional alternative splicing in the human genome?Trends Genet. 2004 Feb;20(2):68-71. doi: 10.1016/j.tig.2003.12.004. Trends Genet. 2004. PMID: 14746986 Review.
Cited by
-
The Impact of Pro-Inflammatory Cytokines on Alternative Splicing Patterns in Human Islets.Diabetes. 2021 Oct 25;71(1):116-27. doi: 10.2337/db20-0847. Online ahead of print. Diabetes. 2021. PMID: 34697029 Free PMC article.
-
ExonImpact: Prioritizing Pathogenic Alternative Splicing Events.Hum Mutat. 2017 Jan;38(1):16-24. doi: 10.1002/humu.23111. Epub 2016 Oct 3. Hum Mutat. 2017. PMID: 27604408 Free PMC article.
-
Sounds of silence: synonymous nucleotides as a key to biological regulation and complexity.Nucleic Acids Res. 2013 Feb 1;41(4):2073-94. doi: 10.1093/nar/gks1205. Epub 2013 Jan 4. Nucleic Acids Res. 2013. PMID: 23293005 Free PMC article. Review.
-
A proteogenomic approach to understand splice isoform functions through sequence and expression-based computational modeling.Brief Bioinform. 2016 Nov;17(6):1024-1031. doi: 10.1093/bib/bbv109. Epub 2016 Jan 6. Brief Bioinform. 2016. PMID: 26740460 Free PMC article.
-
Identification of a new susceptibility variant for multiple sclerosis in OAS1 by population genetics analysis.Hum Genet. 2012 Jan;131(1):87-97. doi: 10.1007/s00439-011-1053-2. Epub 2011 Jul 7. Hum Genet. 2012. PMID: 21735172 Free PMC article.
References
-
- Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. - PubMed
-
- Graveley BR. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 2001;17:100–107. - PubMed
-
- Blencowe BJ. Alternative splicing: new insights from global analyses. Cell. 2006;126:37–47. - PubMed
-
- Stamm S, Ben-Ari S, Rafalska I, Tang Y, Zhang Z, et al. Function of alternative splicing. Gene. 2005;344:1–20. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

