Evolutionary plasticity and innovations in complex metabolic reaction networks
- PMID: 20019795
- PMCID: PMC2785887
- DOI: 10.1371/journal.pcbi.1000613
Evolutionary plasticity and innovations in complex metabolic reaction networks
Abstract
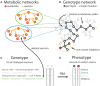
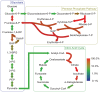
Genome-scale metabolic networks are highly robust to the elimination of enzyme-coding genes. Their structure can evolve rapidly through mutations that eliminate such genes and through horizontal gene transfer that adds new enzyme-coding genes. Using flux balance analysis we study a vast space of metabolic network genotypes and their relationship to metabolic phenotypes, the ability to sustain life in an environment defined by an available spectrum of carbon sources. Two such networks typically differ in most of their reactions and have few essential reactions in common. Our observations suggest that the robustness of the Escherichia coli metabolic network to mutations is typical of networks with the same phenotype. We also demonstrate that networks with the same phenotype form large sets that can be traversed through single mutations, and that single mutations of different genotypes with the same phenotype can yield very different novel phenotypes. This means that the evolutionary plasticity and robustness of metabolic networks facilitates the evolution of new metabolic abilities. Our approach has broad implications for the evolution of metabolic networks, for our understanding of mutational robustness, for the design of antimetabolic drugs, and for metabolic engineering.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Genotype networks, innovation, and robustness in sulfur metabolism.BMC Syst Biol. 2011 Mar 7;5:39. doi: 10.1186/1752-0509-5-39. BMC Syst Biol. 2011. PMID: 21385333 Free PMC article.
-
Genotype networks in metabolic reaction spaces.BMC Syst Biol. 2010 Mar 19;4:30. doi: 10.1186/1752-0509-4-30. BMC Syst Biol. 2010. PMID: 20302636 Free PMC article.
-
Superessential reactions in metabolic networks.Proc Natl Acad Sci U S A. 2012 May 1;109(18):E1121-30. doi: 10.1073/pnas.1113065109. Epub 2012 Apr 16. Proc Natl Acad Sci U S A. 2012. PMID: 22509034 Free PMC article.
-
The molecular origins of evolutionary innovations.Trends Genet. 2011 Oct;27(10):397-410. doi: 10.1016/j.tig.2011.06.002. Epub 2011 Aug 27. Trends Genet. 2011. PMID: 21872964 Review.
-
Genotype networks shed light on evolutionary constraints.Trends Ecol Evol. 2011 Nov;26(11):577-84. doi: 10.1016/j.tree.2011.07.001. Epub 2011 Aug 15. Trends Ecol Evol. 2011. PMID: 21840080 Review.
Cited by
-
Phenotypic innovation through recombination in genome-scale metabolic networks.Proc Biol Sci. 2016 Sep 28;283(1839):20161536. doi: 10.1098/rspb.2016.1536. Proc Biol Sci. 2016. PMID: 27683361 Free PMC article.
-
The translational network for metabolic disease - from protein interaction to disease co-occurrence.BMC Bioinformatics. 2019 Nov 13;20(1):576. doi: 10.1186/s12859-019-3106-9. BMC Bioinformatics. 2019. PMID: 31722666 Free PMC article.
-
Robustness and evolvability.Trends Genet. 2010 Sep;26(9):406-14. doi: 10.1016/j.tig.2010.06.002. Epub 2010 Jul 1. Trends Genet. 2010. PMID: 20598394 Free PMC article. Review.
-
The architecture of an empirical genotype-phenotype map.Evolution. 2018 Jun;72(6):1242-1260. doi: 10.1111/evo.13487. Epub 2018 May 25. Evolution. 2018. PMID: 29676774 Free PMC article.
-
Human genome variation and the concept of genotype networks.PLoS One. 2014 Jun 9;9(6):e99424. doi: 10.1371/journal.pone.0099424. eCollection 2014. PLoS One. 2014. PMID: 24911413 Free PMC article.
References
-
- Li W-H. Massachusetts: Sinauer; 1997. Molecular Evolution.
-
- Edwards JS, Palsson BO. Systems properties of the Haemophilus influenzae Rd metabolic genotype. J Biol Chem. 1999;274:17410–17416. - PubMed
-
- Price ND, Reed JL, Palsson BO. Genome-scale models of microbial cells: evaluating the consequences of constraints. Nature Reviews Microbiology. 2004;2:886–897. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

