Global demethylation of rat chondrosarcoma cells after treatment with 5-aza-2'-deoxycytidine results in increased tumorigenicity
- PMID: 20019818
- PMCID: PMC2790612
- DOI: 10.1371/journal.pone.0008340
Global demethylation of rat chondrosarcoma cells after treatment with 5-aza-2'-deoxycytidine results in increased tumorigenicity
Abstract
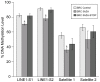
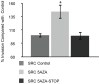
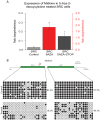
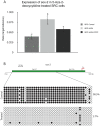
Abnormal patterns of DNA methylation are observed in several types of human cancer. While localized DNA methylation of CpG islands has been associated with gene silencing, the effect that genome-wide loss of methylation has on tumorigenesis is not completely known. To examine its effect on tumorigenesis, we induced DNA demethylation in a rat model of human chondrosarcoma using 5-aza-2-deoxycytidine. Rat specific pyrosequencing assays were utilized to assess the methylation levels in both LINEs and satellite DNA sequences following 5-aza-2-deoxycytidine treatment. Loss of DNA methylation was accompanied by an increase in invasiveness of the rat chondrosarcoma cells, in vitro, as well as by an increase in tumor growth in vivo. Subsequent microarray analysis provided insight into the gene expression changes that result from 5-aza-2-deoxycytidine induced DNA demethylation. In particular, two genes that may function in tumorigenesis, sox-2 and midkine, were expressed at low levels in control cells but upon 5-aza-2-deoxycytidine treatment these genes became overexpressed. Promoter region DNA analysis revealed that these genes were methylated in control cells but became demethylated following 5-aza-2-deoxycytidine treatment. Following withdrawal of 5-aza-2-deoxycytidine, the rat chondrosarcoma cells reestablished global DNA methylation levels that were comparable to that of control cells. Concurrently, invasiveness of the rat chondrosarcoma cells, in vitro, decreased to a level indistinguishable to that of control cells. Taken together these experiments demonstrate that global DNA hypomethylation induced by 5-aza-2-deoxycytidine may promote specific aspects of tumorigenesis in rat chondrosarcoma cells.
Conflict of interest statement
Figures


Similar articles
-
Microarray analysis of gene expression in metastatic gastric cancer cells after incubation with the methylation inhibitor 5-aza-2'-deoxycytidine.Clin Exp Metastasis. 2004;21(5):389-97. doi: 10.1007/s10585-005-1043-7. Clin Exp Metastasis. 2004. PMID: 15672863
-
Epigenetic silencing of SFRP5 promotes the metastasis and invasion of chondrosarcoma by expression inhibition and Wnt signaling pathway activation.Chem Biol Interact. 2018 Dec 25;296:1-8. doi: 10.1016/j.cbi.2018.08.020. Epub 2018 Aug 18. Chem Biol Interact. 2018. PMID: 30125549
-
Genome-wide methylation and expression profiling identifies promoter characteristics affecting demethylation-induced gene up-regulation in melanoma.BMC Med Genomics. 2010 Feb 9;3:4. doi: 10.1186/1755-8794-3-4. BMC Med Genomics. 2010. PMID: 20144234 Free PMC article.
-
Antineoplastic activity of the DNA methyltransferase inhibitor 5-aza-2'-deoxycytidine in anaplastic large cell lymphoma.Biochimie. 2012 Nov;94(11):2297-307. doi: 10.1016/j.biochi.2012.05.029. Epub 2012 Jun 9. Biochimie. 2012. PMID: 22687603 Free PMC article. Review.
-
DNA demethylation and invasive cancer: implications for therapeutics.Br J Pharmacol. 2015 Jun;172(11):2705-15. doi: 10.1111/bph.12885. Epub 2015 Apr 27. Br J Pharmacol. 2015. PMID: 25134627 Free PMC article. Review.
Cited by
-
Writing and rewriting the epigenetic code of cancer cells: from engineered proteins to small molecules.Mol Pharmacol. 2013 Mar;83(3):563-76. doi: 10.1124/mol.112.080697. Epub 2012 Nov 13. Mol Pharmacol. 2013. PMID: 23150486 Free PMC article. Review.
-
Eicosapentaenoic acid demethylates a single CpG that mediates expression of tumor suppressor CCAAT/enhancer-binding protein delta in U937 leukemia cells.J Biol Chem. 2011 Aug 5;286(31):27092-102. doi: 10.1074/jbc.M111.253609. Epub 2011 Jun 9. J Biol Chem. 2011. PMID: 21659508 Free PMC article.
-
Palate morphogenesis: current understanding and future directions.Birth Defects Res C Embryo Today. 2010 Jun;90(2):133-54. doi: 10.1002/bdrc.20180. Birth Defects Res C Embryo Today. 2010. PMID: 20544696 Free PMC article. Review.
-
Epigenetics of Skeletal Diseases.Curr Osteoporos Rep. 2018 Jun;16(3):246-255. doi: 10.1007/s11914-018-0435-y. Curr Osteoporos Rep. 2018. PMID: 29574535 Review.
-
Aging-associated excess formaldehyde leads to spatial memory deficits.Sci Rep. 2013;3:1807. doi: 10.1038/srep01807. Sci Rep. 2013. PMID: 23657727 Free PMC article.
References
-
- Brena RM, Costello JF. Genome-epigenome interactions in cancer. Hum Mol Genet 16 Spec No. 2007;1:R96–105. - PubMed
-
- Mund C, Brueckner B, Lyko F. Reactivation of epigenetically silenced genes by DNA methyltransferase inhibitors: basic concepts and clinical applications. Epigenetics. 2006;1:7–13. - PubMed
-
- Chen L, MacMillan AM, Chang W, Ezaz-Nikpay K, Lane WS, et al. Direct identification of the active-site nucleophile in a DNA (cytosine-5)-methyltransferase. Biochemistry. 1991;30:11018–11025. - PubMed
-
- Mund C, Hackanson B, Stresemann C, Lubbert M, Lyko F. Characterization of DNA demethylation effects induced by 5-Aza-2′-deoxycytidine in patients with myelodysplastic syndrome. Cancer Res. 2005;65:7086–7090. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

