Epigenetic silencing of CRABP2 and MX1 in head and neck tumors
- PMID: 20019841
- PMCID: PMC2794514
- DOI: 10.1593/neo.91110
Epigenetic silencing of CRABP2 and MX1 in head and neck tumors
Abstract
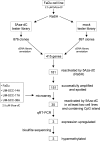
Head and neck squamous cell carcinoma (HNSCC) is a heterogeneous disease affecting the epithelium of the oral cavity, pharynx and larynx. Conditions of most patients are diagnosed at late stages of the disease, and no sensitive and specific predictors of aggressive behavior have been identified yet. Therefore, early detection and prognostic biomarkers are highly desirable for a more rational management of the disease. Hypermethylation of CpG islands is one of the most important epigenetic mechanisms that leads to gene silencing in tumors and has been extensively used for the identification of biomarkers. In this study, we combined rapid subtractive hybridization and microarray analysis in a hierarchical manner to select genes that are putatively reactivated by the demethylating agent 5-aza-2'-deoxycytidine (5Aza-dC) in HNSCC cell lines (FaDu, UM-SCC-14A, UM-SCC-17A, UM-SCC-38A). This combined analysis identified 78 genes, 35 of which were reactivated in at least 2 cell lines and harbored a CpG island at their 5' region. Reactivation of 3 of these 35 genes (CRABP2, MX1, and SLC15A3) was confirmed by quantitative real-time polymerase chain reaction (PCR; fold change, >or=3). Bisulfite sequencing of their CpG islands revealed that they are indeed differentially methylated in the HNSCC cell lines. Using methylation-specific PCR, we detected a higher frequency of CRABP2 (58.1% for region 1) and MX1 (46.3%) hypermethylation in primary HNSCC when compared with lymphocytes from healthy individuals. Finally, absence of the CRABP2 protein was associated with decreased disease-free survival rates, supporting a potential use of CRABP2 expression as a prognostic biomarker for HNSCC patients.
Figures




Similar articles
-
Epigenetic screen of human DNA repair genes identifies aberrant promoter methylation of NEIL1 in head and neck squamous cell carcinoma.Oncogene. 2012 Dec 6;31(49):5108-16. doi: 10.1038/onc.2011.660. Epub 2012 Jan 30. Oncogene. 2012. PMID: 22286769
-
Hypermethylation of the retinoic acid receptor-beta(2) gene in head and neck carcinogenesis.Clin Cancer Res. 2004 Mar 1;10(5):1733-42. doi: 10.1158/1078-0432.ccr-0989-3. Clin Cancer Res. 2004. PMID: 15014026
-
Transporter gene expression in human head and neck squamous cell carcinoma and associated epigenetic regulatory mechanisms.Am J Pathol. 2013 Jan;182(1):234-43. doi: 10.1016/j.ajpath.2012.09.008. Epub 2012 Nov 6. Am J Pathol. 2013. PMID: 23137910
-
Delineating an epigenetic continuum in head and neck cancer.Cancer Lett. 2014 Jan 28;342(2):178-84. doi: 10.1016/j.canlet.2012.02.018. Epub 2012 Mar 1. Cancer Lett. 2014. PMID: 22388100 Free PMC article. Review.
-
DNA Methylation as a Diagnostic, Prognostic, and Predictive Biomarker in Head and Neck Cancer.Int J Mol Sci. 2023 Feb 3;24(3):2996. doi: 10.3390/ijms24032996. Int J Mol Sci. 2023. PMID: 36769317 Free PMC article. Review.
Cited by
-
Epigenetic Regulation in Oral Squamous Cell Carcinoma Microenvironment: A Comprehensive Review.Cancers (Basel). 2023 Nov 27;15(23):5600. doi: 10.3390/cancers15235600. Cancers (Basel). 2023. PMID: 38067304 Free PMC article. Review.
-
FOXD2-AS1 promotes malignant cell behavior in oral squamous cell carcinoma via the miR-378 g/CRABP2 axis.BMC Oral Health. 2024 May 28;24(1):625. doi: 10.1186/s12903-024-04388-2. BMC Oral Health. 2024. PMID: 38807101 Free PMC article.
-
Cellular Retinoic Acid Binding Protein 2 Is Strikingly Downregulated in Human Esophageal Squamous Cell Carcinoma and Functions as a Tumor Suppressor.PLoS One. 2016 Feb 3;11(2):e0148381. doi: 10.1371/journal.pone.0148381. eCollection 2016. PLoS One. 2016. PMID: 26839961 Free PMC article.
-
CRABP-II methylation: a critical determinant of retinoic acid resistance of medulloblastoma cells.Mol Oncol. 2012 Feb;6(1):48-61. doi: 10.1016/j.molonc.2011.11.004. Epub 2011 Nov 25. Mol Oncol. 2012. PMID: 22153617 Free PMC article.
-
Differentially expressed genes in giant cell tumor of bone.Virchows Arch. 2011 Apr;458(4):467-76. doi: 10.1007/s00428-011-1047-4. Epub 2011 Feb 9. Virchows Arch. 2011. PMID: 21305317
References
-
- McMahon S, Chen AY. Head and neck cancer. Cancer Metastasis Rev. 2003;22:21–24. - PubMed
-
- Goldenberg D, Lee J, Koch WM, Kim MM, Trink B, Sidransky D, Moon CS. Habitual risk factors for head and neck cancer. Otolaryngol Head Neck Surg. 2004;131:986–993. - PubMed
-
- Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. - PubMed
-
- Moriniere S. Epidemiology of head and neck cancer [in French] Rev Prat. 2006;56:1637–1641. - PubMed
-
- Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
