An epigenetic signature in peripheral blood predicts active ovarian cancer
- PMID: 20019873
- PMCID: PMC2793425
- DOI: 10.1371/journal.pone.0008274
An epigenetic signature in peripheral blood predicts active ovarian cancer
Abstract
Background: Recent studies have shown that DNA methylation (DNAm) markers in peripheral blood may hold promise as diagnostic or early detection/risk markers for epithelial cancers. However, to date no study has evaluated the diagnostic and predictive potential of such markers in a large case control cohort and on a genome-wide basis.
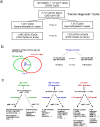
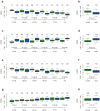
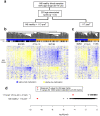
Principal findings: By performing genome-wide DNAm profiling of a large ovarian cancer case control cohort, we here demonstrate that active ovarian cancer has a significant impact on the DNAm pattern in peripheral blood. Specifically, by measuring the methylation levels of over 27,000 CpGs in blood cells from 148 healthy individuals and 113 age-matched pre-treatment ovarian cancer cases, we derive a DNAm signature that can predict the presence of active ovarian cancer in blind test sets with an AUC of 0.8 (95% CI (0.74-0.87)). We further validate our findings in another independent set of 122 post-treatment cases (AUC = 0.76 (0.72-0.81)). In addition, we provide evidence for a significant number of candidate risk or early detection markers for ovarian cancer. Furthermore, by comparing the pattern of methylation with gene expression data from major blood cell types, we here demonstrate that age and cancer elicit common changes in the composition of peripheral blood, with a myeloid skewing that increases with age and which is further aggravated in the presence of ovarian cancer. Finally, we show that most cancer and age associated methylation variability is found at CpGs located outside of CpG islands.
Significance: Our results underscore the potential of DNAm profiling in peripheral blood as a tool for detection or risk-prediction of epithelial cancers, and warrants further in-depth and higher CpG coverage studies to further elucidate this role.
Conflict of interest statement
Figures

References
-
- Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7:21–33. - PubMed
-
- Baylin SB, Ohm JE. Epigenetic gene silencing in cancer - a mechanism for early oncogenic pathway addiction? Nat Rev Cancer. 2006;6:107–116. - PubMed
-
- Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, et al. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39:157–158. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

