Molecular evolution of Drosophila cuticular protein genes
- PMID: 20019874
- PMCID: PMC2793513
- DOI: 10.1371/journal.pone.0008345
Molecular evolution of Drosophila cuticular protein genes
Abstract
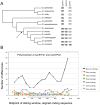
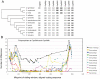
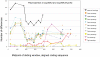
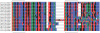
Several multigene families have been described that together encode scores of structural cuticular proteins in Drosophila, although the functional significance of this diversity remains to be explored. Here I investigate the evolutionary histories of several multigene families (CPR, Tweedle, CPLCG, and CPF/CPFL) that vary in age, size, and sequence complexity, using sequenced Drosophila genomes and mosquito outgroups. My objective is to describe the rates and mechanisms of 'cuticle-ome' divergence, in order to identify conserved and rapidly evolving elements. I also investigate potential examples of interlocus gene conversion and concerted evolution within these families during Drosophila evolution. The absolute rate of change in gene number (per million years) is an order of magnitude lower for cuticular protein families within Drosophila than it is among Drosophila and the two mosquito taxa, implying that major transitions in the cuticle proteome have occurred at higher taxonomic levels. Several hotspots of intergenic conversion and/or gene turnover were identified, e.g. some gene pairs have independently undergone intergenic conversion within different lineages. Some gene conversion hotspots were characterized by conversion tracts initiating near nucleotide repeats within coding regions, and similar repeats were found within concertedly evolving cuticular protein genes in Anopheles gambiae. Rates of amino-acid substitution were generally severalfold higher along the branch connecting the Sophophora and Drosophila species groups, and 13 genes have Ka/Ks significantly greater than one along this branch, indicating adaptive divergence. Insect cuticular proteins appear to be a source of adaptive evolution within genera and, at higher taxonomic levels, subject to periods of gene-family expansion and contraction followed by quiescence. However, this relative stasis is belied by hotspots of molecular evolution, particularly concerted evolution, during the diversification of Drosophila. The prominent association between interlocus gene conversion and repeats within the coding sequence of interacting genes suggests that the latter promote strand exchange.
Conflict of interest statement
Figures







References
-
- Neville AC. Berlin: Springer Verlag; 1975. Biology of the Arthropod Cuticle.448
-
- Awolola TS, Oduola OA, Strode C, Koekemoer LL, Brooke B, et al. Evidence of multiple pyrethroid resistance mechanisms in the malaria vector Anopheles gambiae sensu stricto from Nigeria. Trans R Soc Trop Med Hyg 2008 - PubMed
-
- Vontas J, David JP, Nikou D, Hemingway J, Christophides GK, et al. Transcriptional analysis of insecticide resistance in Anopheles stephensi using cross-species microarray hybridization. Insect Mol Biol. 2007;16:315–324. - PubMed
-
- White BJ, Cheng C, Sangare D, Lobo NF, Collins FH, et al. The Population genomics of trans-specific inversion polymorphisms in Anopheles gambiae. Genetics. 2009 DOI: 109.105817. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

