Comparison of the receptor FGFRL1 from sea urchins and humans illustrates evolution of a zinc binding motif in the intracellular domain
- PMID: 20021659
- PMCID: PMC2806250
- DOI: 10.1186/1471-2091-10-33
Comparison of the receptor FGFRL1 from sea urchins and humans illustrates evolution of a zinc binding motif in the intracellular domain
Abstract
Background: FGFRL1, the gene for the fifth member of the fibroblast growth factor receptor (FGFR) family, is found in all vertebrates from fish to man and in the cephalochordate amphioxus. Since it does not occur in more distantly related invertebrates such as insects and nematodes, we have speculated that FGFRL1 might have evolved just before branching of the vertebrate lineage from the other invertebrates (Beyeler and Trueb, 2006).
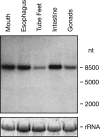
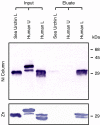
Results: We identified the gene for FGFRL1 also in the sea urchin Strongylocentrotus purpuratus and cloned its mRNA. The deduced amino acid sequence shares 62% sequence similarity with the human protein and shows conservation of all disulfides and N-linked carbohydrate attachment sites. Similar to the human protein, the S. purpuratus protein contains a histidine-rich motif at the C-terminus, but this motif is much shorter than the human counterpart. To analyze the function of the novel motif, recombinant fusion proteins were prepared in a bacterial expression system. The human fusion protein bound to nickel and zinc affinity columns, whereas the sea urchin protein barely interacted with such columns. Direct determination of metal ions by atomic absorption revealed 2.6 mole zinc/mole protein for human FGFRL1 and 1.7 mole zinc/mole protein for sea urchin FGFRL1.
Conclusion: The FGFRL1 gene has evolved much earlier than previously assumed. A comparison of the intracellular domain between sea urchin and human FGFRL1 provides interesting insights into the shaping of a novel zinc binding domain.
Figures
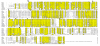

Similar articles
-
Fgfrl1, a fibroblast growth factor receptor-like gene, is found in the cephalochordate Branchiostoma floridae but not in the urochordate Ciona intestinalis.Comp Biochem Physiol B Biochem Mol Biol. 2006 Sep;145(1):43-9. doi: 10.1016/j.cbpb.2006.06.002. Epub 2006 Jun 17. Comp Biochem Physiol B Biochem Mol Biol. 2006. PMID: 16887372
-
Phylogenetic analysis of receptor FgfrL1 shows divergence of the C-terminal end in rodents.Comp Biochem Physiol B Biochem Mol Biol. 2015 Aug;186:43-50. doi: 10.1016/j.cbpb.2015.04.009. Epub 2015 Apr 28. Comp Biochem Physiol B Biochem Mol Biol. 2015. PMID: 25934085
-
Targeted disruption of the intracellular domain of receptor FgfrL1 in mice.PLoS One. 2014 Aug 15;9(8):e105210. doi: 10.1371/journal.pone.0105210. eCollection 2014. PLoS One. 2014. PMID: 25126760 Free PMC article.
-
Biology of FGFRL1, the fifth fibroblast growth factor receptor.Cell Mol Life Sci. 2011 Mar;68(6):951-64. doi: 10.1007/s00018-010-0576-3. Epub 2010 Nov 16. Cell Mol Life Sci. 2011. PMID: 21080029 Free PMC article. Review.
-
The ancestral complement system in sea urchins.Immunol Rev. 2001 Apr;180:16-34. doi: 10.1034/j.1600-065x.2001.1800102.x. Immunol Rev. 2001. PMID: 11414357 Review.
Cited by
-
Remarkable selective constraints on exonic dinucleotide repeats.Evolution. 2014 Sep;68(9):2737-44. doi: 10.1111/evo.12460. Epub 2014 Jul 9. Evolution. 2014. PMID: 24899386 Free PMC article.
-
Fibroblast growth factor receptor like-1 (FGFRL1) interacts with SHP-1 phosphatase at insulin secretory granules and induces beta-cell ERK1/2 protein activation.J Biol Chem. 2013 Jun 14;288(24):17859-70. doi: 10.1074/jbc.M112.440677. Epub 2013 May 2. J Biol Chem. 2013. PMID: 23640895 Free PMC article.
-
Role of FGFRL1 and other FGF signaling proteins in early kidney development.Cell Mol Life Sci. 2013 Jul;70(14):2505-18. doi: 10.1007/s00018-012-1189-9. Epub 2012 Oct 31. Cell Mol Life Sci. 2013. PMID: 23112089 Free PMC article. Review.
-
A net-like structure with pores is observed during cell fusion induced by the receptor FGFRL1.Commun Integr Biol. 2011 May;4(3):287-90. doi: 10.4161/cib.4.3.14892. Commun Integr Biol. 2011. PMID: 21980560 Free PMC article.
-
miR-210 promotes human osteosarcoma cell migration and invasion by targeting FGFRL1.Oncol Lett. 2018 Aug;16(2):2229-2236. doi: 10.3892/ol.2018.8939. Epub 2018 Jun 11. Oncol Lett. 2018. PMID: 30008923 Free PMC article.
References
-
- Kim I, Moon S-O, Yu K-H, Kim U-H, Koh GY. A novel fibroblast growth factor receptor-5 preferentially expressed in the pancreas. Biochim Biophys Acta. 2001;1518(1-2):152–156. - PubMed
-
- Trueb B, Taeschler S. Expression of FGFRL1, a novel fibroblast growth factor receptor, during embryonic development. Int J Mol Med. . 2006;17(4):617–620. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

