Gene expression relationship between prostate cancer cells of Gleason 3, 4 and normal epithelial cells as revealed by cell type-specific transcriptomes
- PMID: 20021671
- PMCID: PMC2809079
- DOI: 10.1186/1471-2407-9-452
Gene expression relationship between prostate cancer cells of Gleason 3, 4 and normal epithelial cells as revealed by cell type-specific transcriptomes
Abstract
Background: Prostate cancer cells in primary tumors have been typed CD10-/CD13-/CD24hi/CD26+/CD38lo/CD44-/CD104-. This CD phenotype suggests a lineage relationship between cancer cells and luminal cells. The Gleason grade of tumors is a descriptive of tumor glandular differentiation. Higher Gleason scores are associated with treatment failure.
Methods: CD26+ cancer cells were isolated from Gleason 3+3 (G3) and Gleason 4+4 (G4) tumors by cell sorting, and their gene expression or transcriptome was determined by Affymetrix DNA array analysis. Dataset analysis was used to determine gene expression similarities and differences between G3 and G4 as well as to prostate cancer cell lines and histologically normal prostate luminal cells.
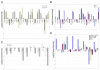
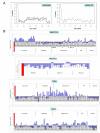
Results: The G3 and G4 transcriptomes were compared to those of prostatic cell types of non-cancer, which included luminal, basal, stromal fibromuscular, and endothelial. A principal components analysis of the various transcriptome datasets indicated a closer relationship between luminal and G3 than luminal and G4. Dataset comparison also showed that the cancer transcriptomes differed substantially from those of prostate cancer cell lines.
Conclusions: Genes differentially expressed in cancer are potential biomarkers for cancer detection, and those differentially expressed between G3 and G4 are potential biomarkers for disease stratification given that G4 cancer is associated with poor outcomes. Differentially expressed genes likely contribute to the prostate cancer phenotype and constitute the signatures of these particular cancer cell types.
Figures




References
-
- True L, Coleman I, Hawley S, Huang CY, Gifford D, Coleman R, Beer TM, Gelmann E, Datta M, Mostaghel E, Knudsen B, Lange P, Vessella R, Lin D, Hood L, Nelson PS. A molecular correlate to the Gleason grading system for prostate adenocarcinoma. Proc Natl Acad Sci USA. 2006;103(29):10991–10996. doi: 10.1073/pnas.0603678103. - DOI - PMC - PubMed
-
- Reis EM, Nakaya HI, Louro R, Canavez FC, Flatschart ÁV, Almeida GT, Egidio CM, Paquola AC, Machado AA, Festa F, Yamamoto D, Alvarenga R, da Silva CC, Brito GC, Simon SD, Moreira-Filho CA, Leite KR, Camara-Lopes LH, Campos FS, Gimba E, Vignal GM, El-Dorry H, Sogayar MC, Barcinski MA, da Silva AM, Verjovski-Almeida S. Antisense intronic non-coding RNA levels correlate to the degree of tumor differentiation in prostate cancer. Oncogene. 2004;23(39):6684–6692. doi: 10.1038/sj.onc.1207880. - DOI - PubMed
-
- Bettuzzi S, Scaltriti M, Caporali A, Brausi M, D'Arca D, Astancolle S, Davalli P, Corti A. Successful prediction of prostate cancer recurrence by gene profiling in combination with clinical data: a 5-year follow-up study. Cancer Res. 2003;63(13):3469–3472. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

