Her4 and Her2/neu tyrosine kinase domains dimerize and activate in a reconstituted in vitro system
- PMID: 20022944
- PMCID: PMC2844153
- DOI: 10.1074/jbc.M109.096032
Her4 and Her2/neu tyrosine kinase domains dimerize and activate in a reconstituted in vitro system
Abstract
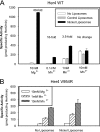
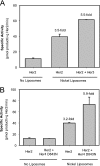
Her4 (ErbB-4) and Her2/neu (ErbB-2) are receptor-tyrosine kinases belonging to the epidermal growth factor receptor (EGFR) family. Crystal structures of EGFR and Her4 kinase domains demonstrate kinase dimerization and activation through an allosteric mechanism. The kinase domains form an asymmetric dimer, where the C-lobe surface of one monomer contacts the N-lobe of the other monomer. EGFR kinase dimerization and activation in vitro was previously reported using a nickel-chelating lipid-liposome system, and we now apply this system to all other members of the EGFR family. Polyhistidine-tagged Her4, Her2/neu, and Her3 kinase domains are bound to these nickel-liposomes and are brought to high local concentration, mimicking what happens to full-length receptors in vivo following ligand binding. Addition of nickel-liposomes to Her4 kinase domain results in 40-fold activation in kinase activity and marked enhancement of C-terminal tail autophosphorylation. Activation of Her4 shows a sigmoidal dependence on kinase concentration, consistent with a cooperative process requiring kinase dimerization. Her2/neu kinase activity is also activated by nickel-liposomes, and is increased further by heterodimerization with Her3 or Her4. The ability of Her3 and Her4 to heterodimerize and activate other family members is studied in vitro. Her3 kinase domain readily activates Her2/neu but is a poor activator of Her4, which differs from the prediction made by the asymmetric dimer model. Mutation of Her3 residues (952)ENI(954) to the corresponding sequence in Her4 enhanced the ability of Her3 to activate Her4, demonstrating that sequence differences on the C-lobe surface influence the heterodimerization and activation of ErbB kinase domains.
Figures



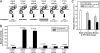

Similar articles
-
HER3 intracellular domains play a crucial role in HER3/HER2 dimerization and activation of downstream signaling pathways.Protein Cell. 2012 Oct;3(10):781-9. doi: 10.1007/s13238-012-2065-y. Epub 2012 Sep 15. Protein Cell. 2012. PMID: 22983903 Free PMC article.
-
Structural analysis of the catalytically inactive kinase domain of the human EGF receptor 3.Proc Natl Acad Sci U S A. 2009 Dec 22;106(51):21608-13. doi: 10.1073/pnas.0912101106. Epub 2009 Dec 9. Proc Natl Acad Sci U S A. 2009. PMID: 20007378 Free PMC article.
-
Structural analysis of the EGFR/HER3 heterodimer reveals the molecular basis for activating HER3 mutations.Sci Signal. 2014 Dec 2;7(354):ra114. doi: 10.1126/scisignal.2005786. Sci Signal. 2014. PMID: 25468994 Free PMC article.
-
ErbB Receptors and Cancer.Methods Mol Biol. 2017;1652:3-35. doi: 10.1007/978-1-4939-7219-7_1. Methods Mol Biol. 2017. PMID: 28791631 Review.
-
Heterodimerization and functional interaction between EGF receptor family members: a new signaling paradigm with implications for breast cancer research.Breast Cancer Res Treat. 1995 Jul;35(1):115-32. doi: 10.1007/BF00694752. Breast Cancer Res Treat. 1995. PMID: 7612898 Review.
Cited by
-
Phosphoproteomics of collagen receptor networks reveals SHP-2 phosphorylation downstream of wild-type DDR2 and its lung cancer mutants.Biochem J. 2013 Sep 15;454(3):501-13. doi: 10.1042/BJ20121750. Biochem J. 2013. PMID: 23822953 Free PMC article.
-
Expression of HER2 in Breast Cancer Promotes a Massive Reorganization of Gene Activity and Suggests a Role for Epigenetic Regulation.J Data Mining Genomics Proteomics. 2012;3:e102. doi: 10.4172/2153-0602.1000e102. J Data Mining Genomics Proteomics. 2012. PMID: 24009986 Free PMC article. No abstract available.
-
A highly efficient peptide substrate for EGFR activates the kinase by inducing aggregation.Biochem J. 2013 Aug 1;453(3):337-44. doi: 10.1042/BJ20130537. Biochem J. 2013. PMID: 23734957 Free PMC article.
-
Carboxyl group footprinting mass spectrometry and molecular dynamics identify key interactions in the HER2-HER3 receptor tyrosine kinase interface.J Biol Chem. 2013 Aug 30;288(35):25254-25264. doi: 10.1074/jbc.M113.474882. Epub 2013 Jul 10. J Biol Chem. 2013. PMID: 23843458 Free PMC article.
-
Single-molecule functional anatomy of endogenous HER2-HER3 heterodimers.Elife. 2020 Apr 8;9:e53934. doi: 10.7554/eLife.53934. Elife. 2020. PMID: 32267234 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

