Automated detection of inaccurate and imprecise transitions in peptide quantification by multiple reaction monitoring mass spectrometry
- PMID: 20022980
- PMCID: PMC2851178
- DOI: 10.1373/clinchem.2009.138420
Automated detection of inaccurate and imprecise transitions in peptide quantification by multiple reaction monitoring mass spectrometry
Abstract
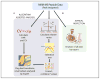
Background: Multiple reaction monitoring mass spectrometry (MRM-MS) of peptides with stable isotope-labeled internal standards (SISs) is increasingly being used to develop quantitative assays for proteins in complex biological matrices. These assays can be highly precise and quantitative, but the frequent occurrence of interferences requires that MRM-MS data be manually reviewed, a time-intensive process subject to human error. We developed an algorithm that identifies inaccurate transition data based on the presence of interfering signal or inconsistent recovery among replicate samples.
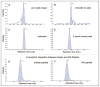
Methods: The algorithm objectively evaluates MRM-MS data with 2 orthogonal approaches. First, it compares the relative product ion intensities of the analyte peptide to those of the SIS peptide and uses a t-test to determine if they are significantly different. A CV is then calculated from the ratio of the analyte peak area to the SIS peak area from the sample replicates.
Results: The algorithm identified problematic transitions and achieved accuracies of 94%-100%, with a sensitivity and specificity of 83%-100% for correct identification of errant transitions. The algorithm was robust when challenged with multiple types of interferences and problematic transitions.
Conclusions: This algorithm for automated detection of inaccurate and imprecise transitions (AuDIT) in MRM-MS data reduces the time required for manual and subjective inspection of data, improves the overall accuracy of data analysis, and is easily implemented into the standard data-analysis work flow. AuDIT currently works with results exported from MRM-MS data-processing software packages and may be implemented as an analysis tool within such software.
Conflict of interest statement
Figures



References
-
- Yost RA, Enke CG. Triple quadrupole mass spectrometry for direct mixture analysis and structure elucidation. Anal Chem. 1979;51:1251–64. - PubMed
-
- Brumley WC, Sphon JA. Regulatory mass spectrometry. Biomed Mass Spectrom. 1981;8:390–6. - PubMed
-
- Sphon JA. Use of mass spectrometry for confirmation of animal drug residues. J Assoc Off Anal Chem. 1978;61:1247–52. - PubMed
-
- Vargo JD. Determination of sulfonic acid degradates of chloroacetanilide and chloroacetamide herbicides in groundwater by LC/MS/MS. Anal Chem. 1998;70:2699–703. - PubMed
-
- Draisci R, Palleschi L, Ferretti E, Lucentini L, Cammarata P. Quantitation of anabolic hormones and their metabolites in bovine serum and urine by liquid chromatography-tandem mass spectrometry. J Chromatogr A. 2000;870:511–22. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

