A novel pheromone quorum-sensing system controls the development of natural competence in Streptococcus thermophilus and Streptococcus salivarius
- PMID: 20023010
- PMCID: PMC2820839
- DOI: 10.1128/JB.01251-09
A novel pheromone quorum-sensing system controls the development of natural competence in Streptococcus thermophilus and Streptococcus salivarius
Abstract
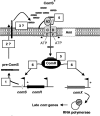
In streptococcal species, the key step of competence development is the transcriptional induction of comX, which encodes the alternative sigma factor sigma(X), which positively regulates genes necessary for DNA transformation. In Streptococcus species belonging to the mitis and mutans groups, induction of comX relies on the activation of a three-component system consisting of a secreted pheromone, a histidine kinase, and a response regulator. In Streptococcus thermophilus, a species belonging to the salivarius group, the oligopeptide transporter Ami is essential for comX expression under competence-inducing conditions. This suggests a different regulation pathway of competence based on the production and reimportation of a signal peptide. The objective of our work was to identify the main actors involved in the early steps of comX induction in S. thermophilus LMD-9. Using a transcriptomic approach, four highly induced early competence operons were identified. Among them, we found a Rgg-like regulator (Ster_0316) associated with a nonannotated gene encoding a 24-amino-acid hydrophobic peptide (Shp0316). Through genetic deletions, we showed that these two genes are essential for comX induction. Moreover, addition to the medium of synthetic peptides derived from the C-terminal part of Shp0316 restored comX induction and transformation of a Shp0316-deficient strain. These peptides also induced competence in S. thermophilus and Streptococcus salivarius strains that are poorly transformable or not transformable. Altogether, our results show that Ster_0316 and Shp0316, renamed ComRS, are the two members of a novel quorum-sensing system responsible for comX induction in species from the salivarius group, which differs from the classical phosphorelay three-component system identified previously in streptococci.
Figures


References
-
- Alloing, G., B. Martin, C. Granadel, and J. P. Claverys. 1998. Development of competence in Streptococcus pneumonaie: pheromone autoinduction and control of quorum sensing by the oligopeptide permease. Mol. Microbiol. 29:75-83. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

