Characterization of a glutamate transporter operon, glnQHMP, in Streptococcus mutans and its role in acid tolerance
- PMID: 20023025
- PMCID: PMC2812961
- DOI: 10.1128/JB.01169-09
Characterization of a glutamate transporter operon, glnQHMP, in Streptococcus mutans and its role in acid tolerance
Abstract
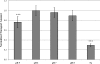
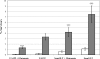
Glutamate contributes to the acid tolerance response (ATR) of many Gram-negative and Gram-positive bacteria, but its role in the ATR of the oral bacterium Streptococcus mutans is unknown. This study describes the discovery and characterization of a glutamate transporter operon designated glnQHMP (Smu.1519 to Smu.1522) and investigates its potential role in acid tolerance. Deletion of glnQHMP resulted in a 95% reduction in transport of radiolabeled glutamate compared to the wild-type UA159 strain. The addition of glutamate to metabolizing UA159 cells resulted in an increased production of acidic end products, whereas the glnQHMP mutant produced less lactic acid than UA159, suggesting a link between glutamate metabolism and acid production and possible acid tolerance. To investigate this possibility, we conducted a microarray analysis with glutamate and under pH 5.5 and pH 7.5 conditions which showed that expression of the glnQHMP operon was downregulated by both glutamate and mild acid. We also measured the growth kinetics of UA159 and its glnQHMP-negative derivative at pH 5.5 and found that the mutant doubled at a much slower rate than the parent strain but survived at pH 3.5 significantly better than the wild type. Taken together, these findings support the involvement of the glutamate transporter operon glnQHMP in the acid tolerance response in S. mutans.
Figures





Similar articles
-
Role of GlnR in acid-mediated repression of genes encoding proteins involved in glutamine and glutamate metabolism in Streptococcus mutans.Appl Environ Microbiol. 2010 Apr;76(8):2478-86. doi: 10.1128/AEM.02622-09. Epub 2010 Feb 19. Appl Environ Microbiol. 2010. PMID: 20173059 Free PMC article.
-
Characterization of the Streptococcus mutans SMU.1703c-SMU.1702c Operon Reveals Its Role in Riboflavin Import and Response to Acid Stress.J Bacteriol. 2020 Dec 18;203(2):e00293-20. doi: 10.1128/JB.00293-20. Print 2020 Dec 18. J Bacteriol. 2020. PMID: 33077636 Free PMC article.
-
The copYAZ Operon Functions in Copper Efflux, Biofilm Formation, Genetic Transformation, and Stress Tolerance in Streptococcus mutans.J Bacteriol. 2015 Aug 1;197(15):2545-57. doi: 10.1128/JB.02433-14. Epub 2015 May 26. J Bacteriol. 2015. PMID: 26013484 Free PMC article.
-
Dlt operon regulates physiological function and cariogenic virulence in Streptococcus mutans.Future Microbiol. 2023 Mar;18:225-233. doi: 10.2217/fmb-2022-0165. Epub 2023 Apr 25. Future Microbiol. 2023. PMID: 37097048 Review.
-
Acid tolerance mechanisms utilized by Streptococcus mutans.Future Microbiol. 2010 Mar;5(3):403-17. doi: 10.2217/fmb.09.129. Future Microbiol. 2010. PMID: 20210551 Free PMC article. Review.
Cited by
-
Effects of Copper and pH on the Growth and Physiology of Desmodesmus sp. AARLG074.Metabolites. 2019 Apr 30;9(5):84. doi: 10.3390/metabo9050084. Metabolites. 2019. PMID: 31052259 Free PMC article.
-
Transcriptional profile of glucose-shocked and acid-adapted strains of Streptococcus mutans.Mol Oral Microbiol. 2015 Dec;30(6):496-517. doi: 10.1111/omi.12110. Epub 2015 Jul 2. Mol Oral Microbiol. 2015. PMID: 26042838 Free PMC article.
-
Exploration of Survival Traits, Probiotic Determinants, Host Interactions, and Functional Evolution of Bifidobacterial Genomes Using Comparative Genomics.Genes (Basel). 2018 Oct 1;9(10):477. doi: 10.3390/genes9100477. Genes (Basel). 2018. PMID: 30275399 Free PMC article.
-
Streptococcus mutans glutamate binding protein (GlnH) as antigen target for a mucosal anti-caries vaccine.Braz J Microbiol. 2022 Dec;53(4):1941-1949. doi: 10.1007/s42770-022-00823-0. Epub 2022 Sep 13. Braz J Microbiol. 2022. PMID: 36098933 Free PMC article.
-
Loss of NADH Oxidase Activity in Streptococcus mutans Leads to Rex-Mediated Overcompensation in NAD+ Regeneration by Lactate Dehydrogenase.J Bacteriol. 2015 Dec;197(23):3645-57. doi: 10.1128/JB.00383-15. Epub 2015 Sep 8. J Bacteriol. 2015. PMID: 26350138 Free PMC article.
References
-
- Alekshun, M. N., S. B. Levy, T. R. Mealy, B. A. Seaton, and J. F. Head. 2001. The crystal structure of MarR, a regulator of multiple antibiotic resistance, at 2.3 Å resolution. Nat. Struct. Biol. 8:710-714. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

