Functional role of Bradyrhizobium japonicum trehalose biosynthesis and metabolism genes during physiological stress and nodulation
- PMID: 20023090
- PMCID: PMC2820964
- DOI: 10.1128/AEM.02483-09
Functional role of Bradyrhizobium japonicum trehalose biosynthesis and metabolism genes during physiological stress and nodulation
Abstract
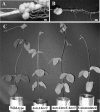
Trehalose, a disaccharide accumulated by many microorganisms, acts as a protectant during periods of physiological stress, such as salinity and desiccation. Previous studies reported that the trehalose biosynthetic genes (otsA, treS, and treY) in Bradyrhizobium japonicum were induced by salinity and desiccation stresses. Functional mutational analyses indicated that disruption of otsA decreased trehalose accumulation in cells and that an otsA treY double mutant accumulated an extremely low level of trehalose. In contrast, trehalose accumulated to a greater extent in a treS mutant, and maltose levels decreased relative to that seen with the wild-type strain. Mutant strains lacking the OtsA pathway, including the single, double, and triple DeltaotsA, DeltaotsA DeltatreS and DeltaotsA DeltatreY, and DeltaotsA DeltatreS DeltatreY mutants, were inhibited for growth on 60 mM NaCl. While mutants lacking functional OtsAB and TreYZ pathways failed to grow on complex medium containing 60 mM NaCl, there was no difference in the viability of the double mutant strain when cells were grown under conditions of desiccation stress. In contrast, mutants lacking a functional TreS pathway were less tolerant of desiccation stress than the wild-type strain. Soybean plants inoculated with mutants lacking the OtsAB and TreYZ pathways produced fewer mature nodules and a greater number of immature nodules relative to those produced by the wild-type strain. Taken together, results of these studies indicate that stress-induced trehalose biosynthesis in B. japonicum is due mainly to the OtsAB pathway and that the TreS pathway is likely involved in the degradation of trehalose to maltose. Trehalose accumulation in B. japonicum enhances survival under conditions of salinity stress and plays a role in the development of symbiotic nitrogen-fixing root nodules on soybean plants.
Figures




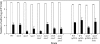

Similar articles
-
Bradyrhizobium diazoefficiens Requires Chemical Chaperones To Cope with Osmotic Stress during Soybean Infection.mBio. 2021 Mar 30;12(2):e00390-21. doi: 10.1128/mBio.00390-21. mBio. 2021. PMID: 33785618 Free PMC article.
-
Trehalose biosynthesis in Rhizobium leguminosarum bv. trifolii and its role in desiccation tolerance.Appl Environ Microbiol. 2007 Jun;73(12):3984-92. doi: 10.1128/AEM.00412-07. Epub 2007 Apr 20. Appl Environ Microbiol. 2007. PMID: 17449695 Free PMC article.
-
Transcriptional and physiological responses of Bradyrhizobium japonicum to desiccation-induced stress.J Bacteriol. 2007 Oct;189(19):6751-62. doi: 10.1128/JB.00533-07. Epub 2007 Jul 27. J Bacteriol. 2007. PMID: 17660288 Free PMC article.
-
Trends in bacterial trehalose metabolism and significant nodes of metabolic pathway in the direction of trehalose accumulation.Microb Biotechnol. 2013 Sep;6(5):493-502. doi: 10.1111/1751-7915.12029. Epub 2013 Jan 10. Microb Biotechnol. 2013. PMID: 23302511 Free PMC article. Review.
-
Why can't vertebrates synthesize trehalose?J Mol Evol. 2014 Oct;79(3-4):111-6. doi: 10.1007/s00239-014-9645-9. Epub 2014 Sep 18. J Mol Evol. 2014. PMID: 25230776 Review.
Cited by
-
A comprehensive genomic, transcriptomic and proteomic analysis of a hyperosmotic stress sensitive α-proteobacterium.BMC Microbiol. 2015 Mar 26;15:71. doi: 10.1186/s12866-015-0404-x. BMC Microbiol. 2015. PMID: 25879753 Free PMC article.
-
Light Modulates Metabolic Pathways and Other Novel Physiological Traits in the Human Pathogen Acinetobacter baumannii.J Bacteriol. 2017 Apr 25;199(10):e00011-17. doi: 10.1128/JB.00011-17. Print 2017 May 15. J Bacteriol. 2017. PMID: 28289081 Free PMC article.
-
Synergistic co-metabolism enhancing the crude oil degradation by Acinetobacter oleivorans DR1 and its metabolic potential.Microbiol Spectr. 2025 Jul;13(7):e0302324. doi: 10.1128/spectrum.03023-24. Epub 2025 May 21. Microbiol Spectr. 2025. PMID: 40396791 Free PMC article.
-
Biosynthesis of compatible solutes in rhizobial strains isolated from Phaseolus vulgaris nodules in Tunisian fields.BMC Microbiol. 2010 Jul 16;10:192. doi: 10.1186/1471-2180-10-192. BMC Microbiol. 2010. PMID: 20633304 Free PMC article.
-
Trehalose and α-glucan mediate distinct abiotic stress responses in Pseudomonas aeruginosa.PLoS Genet. 2021 Apr 19;17(4):e1009524. doi: 10.1371/journal.pgen.1009524. eCollection 2021 Apr. PLoS Genet. 2021. PMID: 33872310 Free PMC article.
References
-
- Ampomah, O. Y., J. B. Jensen, and T. V. Bhuvaneswari. 2008. Lack of trehalose catabolism in Sinorhizobium species increases their nodulation competitiveness on certain host genotypes. New Phytol. 179:495-504. - PubMed
-
- Chang, W. S., W. L. Franck, E. Cytryn, S. Jeong, T. Joshi, D. W. Emerich, M. J. Sadowsky, D. Xu, and G. Stacey. 2007. An oligonucleotide microarray resource for transcriptional profiling of Bradyrhizobium japonicum. Mol. Plant Microbe Interact. 20:1298-1307. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

