Dissecting the regulatory switches of development: lessons from enhancer evolution in Drosophila
- PMID: 20023155
- PMCID: PMC2796927
- DOI: 10.1242/dev.036160
Dissecting the regulatory switches of development: lessons from enhancer evolution in Drosophila
Abstract
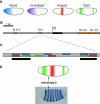
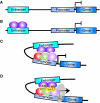
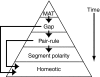
Cis-regulatory modules are non-protein-coding regions of DNA essential for the control of gene expression. One class of regulatory modules is embryonic enhancers, which drive gene expression during development as a result of transcription factor protein binding at the enhancer sequences. Recent comparative studies have begun to investigate the evolution of the sequence architecture within enhancers. These analyses are illuminating the way that developmental biologists think about enhancers by revealing their molecular mechanism of function.
Figures



Similar articles
-
Genome-scale functional characterization of Drosophila developmental enhancers in vivo.Nature. 2014 Aug 7;512(7512):91-5. doi: 10.1038/nature13395. Epub 2014 Jun 1. Nature. 2014. PMID: 24896182
-
Extraction of functional binding sites from unique regulatory regions: the Drosophila early developmental enhancers.Genome Res. 2002 Mar;12(3):470-81. doi: 10.1101/gr.212502. Genome Res. 2002. PMID: 11875036 Free PMC article.
-
Evolution of developmental genes: molecular microevolution of enhancer sequences at the Ubx locus in Drosophila and its impact on developmental phenotypes.Mol Biol Evol. 2004 Feb;21(2):348-63. doi: 10.1093/molbev/msh025. Epub 2003 Dec 5. Mol Biol Evol. 2004. PMID: 14660693
-
Dynamic patterning by morphogens illuminated by cis-regulatory studies.Development. 2021 Jan 20;148(2):dev196113. doi: 10.1242/dev.196113. Development. 2021. PMID: 33472851 Free PMC article. Review.
-
Tempo and mode in evolution of transcriptional regulation.PLoS Genet. 2012 Jan;8(1):e1002432. doi: 10.1371/journal.pgen.1002432. Epub 2012 Jan 19. PLoS Genet. 2012. PMID: 22291600 Free PMC article. Review.
Cited by
-
Intronic cis-regulatory modules mediate tissue-specific and microbial control of angptl4/fiaf transcription.PLoS Genet. 2012;8(3):e1002585. doi: 10.1371/journal.pgen.1002585. Epub 2012 Mar 29. PLoS Genet. 2012. PMID: 22479192 Free PMC article.
-
Color-Patterns to Architecture Conversion through Conditional Generative Adversarial Networks.Biomimetics (Basel). 2021 Feb 17;6(1):16. doi: 10.3390/biomimetics6010016. Biomimetics (Basel). 2021. PMID: 33671287 Free PMC article.
-
Role of architecture in the function and specificity of two Notch-regulated transcriptional enhancer modules.PLoS Genet. 2012 Jul;8(7):e1002796. doi: 10.1371/journal.pgen.1002796. Epub 2012 Jul 5. PLoS Genet. 2012. PMID: 22792075 Free PMC article.
-
In vivo mapping of mutagenesis sensitivity of human enhancers.Nature. 2025 Jul;643(8072):839-846. doi: 10.1038/s41586-025-09182-w. Epub 2025 Jun 18. Nature. 2025. PMID: 40533554
-
Mutagenesis Sensitivity Mapping of Human Enhancers In Vivo.bioRxiv [Preprint]. 2024 Sep 8:2024.09.06.611737. doi: 10.1101/2024.09.06.611737. bioRxiv. 2024. Update in: Nature. 2025 Jul;643(8072):839-846. doi: 10.1038/s41586-025-09182-w. PMID: 39282388 Free PMC article. Updated. Preprint.
References
-
- Andrioli L. P., Vasisht V., Theodosopoulou E., Oberstein A., Small S. (2002). Anterior repression of a Drosophila stripe enhancer requires three position-specific mechanisms. Development 129, 4931-4940 - PubMed
-
- Arnosti D. N., Barolo S., Levine M., Small S. (1996). The eve stripe 2 enhancer employs multiple modes of transcriptional synergy. Development 122, 205-214 - PubMed
-
- Banerji J., Rusconi S., Schaffner W. (1981). Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell 27, 299-308 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

