Dissecting the regulatory switches of development: lessons from enhancer evolution in Drosophila
- PMID: 20023155
- PMCID: PMC2796927
- DOI: 10.1242/dev.036160
Dissecting the regulatory switches of development: lessons from enhancer evolution in Drosophila
Abstract
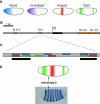
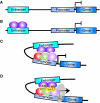
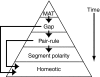
Cis-regulatory modules are non-protein-coding regions of DNA essential for the control of gene expression. One class of regulatory modules is embryonic enhancers, which drive gene expression during development as a result of transcription factor protein binding at the enhancer sequences. Recent comparative studies have begun to investigate the evolution of the sequence architecture within enhancers. These analyses are illuminating the way that developmental biologists think about enhancers by revealing their molecular mechanism of function.
Figures



References
-
- Andrioli L. P., Vasisht V., Theodosopoulou E., Oberstein A., Small S. (2002). Anterior repression of a Drosophila stripe enhancer requires three position-specific mechanisms. Development 129, 4931-4940 - PubMed
-
- Arnosti D. N., Barolo S., Levine M., Small S. (1996). The eve stripe 2 enhancer employs multiple modes of transcriptional synergy. Development 122, 205-214 - PubMed
-
- Banerji J., Rusconi S., Schaffner W. (1981). Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell 27, 299-308 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

